
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
AZD6482 PI3K inhibitor
Cat.No.S1462
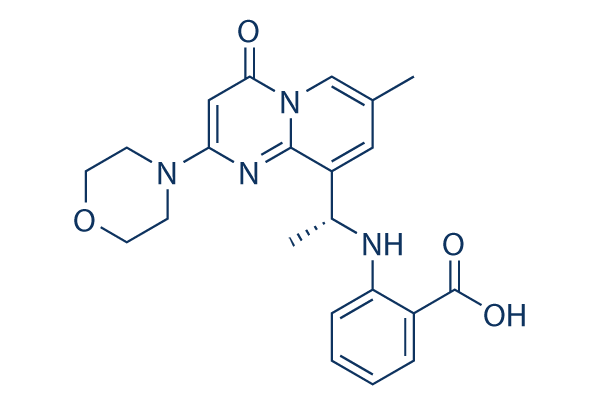
Chemical Structure
Molecular Weight: 408.45
Quality Control
Batch:
Purity:
99.92%
99.92
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human RXF393 cell | Growth inhibition assay | Inhibition of human RXF393 cell growth in a cell viability assay, IC50=0.01154 μM | ||||
| human SW982 cell | Growth inhibition assay | Inhibition of human SW982 cell growth in a cell viability assay, IC50=0.03584 μM | ||||
| human MAD-MB-468 cells | Function assay | Inhibition of PI3Kbeta in human MAD-MB-468 cells assessed as inhibition of Ser473 Akt phosphorylation by cellular potency assay, IC50=0.04 μM | ||||
| human KURAMOCHI cell | Growth inhibition assay | Inhibition of human KURAMOCHI cell growth in a cell viability assay, IC50=0.1051 μM | ||||
| human A498 cell | Growth inhibition assay | Inhibition of human A498 cell growth in a cell viability assay, IC50=0.11795 μM | ||||
| human YT cell | Growth inhibition assay | Inhibition of human YT cell growth in a cell viability assay, IC50=0.12066 μM | ||||
| human VMRC-RCZ cell | Growth inhibition assay | Inhibition of human VMRC-RCZ cell growth in a cell viability assay, IC50=0.14898 μM | ||||
| human MDA-MB-415 cell | Growth inhibition assay | Inhibition of human MDA-MB-415 cell growth in a cell viability assay, IC50=0.16732 μM | ||||
| human J82 cell | Growth inhibition assay | Inhibition of human J82 cell growth in a cell viability assay, IC50=0.47779 μM | ||||
| human SW1710 cell | Growth inhibition assay | Inhibition of human SW1710 cell growth in a cell viability assay, IC50=0.5292 μM | ||||
| human T47D cell | Growth inhibition assay | Inhibition of human T47D cell growth in a cell viability assay, IC50=0.5327 μM | ||||
| human SU-DHL-1 cell | Growth inhibition assay | Inhibition of human SU-DHL-1 cell growth in a cell viability assay, IC50=0.56813 μM | ||||
| human BT-20 cell | Growth inhibition assay | Inhibition of human BT-20 cell growth in a cell viability assay, IC50=0.59222 μM | ||||
| human OS-RC-2 cell | Growth inhibition assay | Inhibition of human OS-RC-2 cell growth in a cell viability assay, IC50=0.62388 μM | ||||
| human RPMI-6666 cell | Growth inhibition assay | Inhibition of human RPMI-6666 cell growth in a cell viability assay, IC50=0.64379 μM | ||||
| human OVCAR-3 cell | Growth inhibition assay | Inhibition of human OVCAR-3 cell growth in a cell viability assay, IC50=0.67946 μM | ||||
| human NALM-6 cell | Growth inhibition assay | Inhibition of human NALM-6 cell growth in a cell viability assay, IC50=0.70465 μM | ||||
| human RS4-11 cell | Growth inhibition assay | Inhibition of human RS4-11 cell growth in a cell viability assay, IC50=0.73806 μM | ||||
| human SK-MES-1 cell | Growth inhibition assay | Inhibition of human SK-MES-1 cell growth in a cell viability assay, IC50=0.78967 μM | ||||
| human ES7 cell | Growth inhibition assay | Inhibition of human ES7 cell growth in a cell viability assay, IC50=0.81286 μM | ||||
| human NCI-H2342 cell | Growth inhibition assay | Inhibition of human NCI-H2342 cell growth in a cell viability assay, IC50=0.84294 μM | ||||
| human SCC-9 cell | Growth inhibition assay | Inhibition of human SCC-9 cell growth in a cell viability assay, IC50=0.8577 μM | ||||
| human EW-7 cell | Growth inhibition assay | Inhibition of human EW-7 cell growth in a cell viability assay, IC50=0.86356 μM | ||||
| human HH cell | Growth inhibition assay | Inhibition of human HH cell growth in a cell viability assay, IC50=0.96222 μM | ||||
| human A427 cell | Growth inhibition assay | Inhibition of human A427 cell growth in a cell viability assay, IC50=0.99505 μM | ||||
| human Daudi cell | Growth inhibition assay | Inhibition of human Daudi cell growth in a cell viability assay, IC50=1.06888 μM | ||||
| human P30-OHK cell | Growth inhibition assay | Inhibition of human P30-OHK cell growth in a cell viability assay, IC50=1.07138 μM | ||||
| human HGC-27 cell | Growth inhibition assay | Inhibition of human HGC-27 cell growth in a cell viability assay, IC50=1.1816 μM | ||||
| human CAMA-1 cell | Growth inhibition assay | Inhibition of human CAMA-1 cell growth in a cell viability assay, IC50=1.2001 μM | ||||
| human SK-OV-3 cell | Growth inhibition assay | Inhibition of human SK-OV-3 cell growth in a cell viability assay, IC50=1.20444 μM | ||||
| human NCI-H1048 cell | Growth inhibition assay | Inhibition of human NCI-H1048 cell growth in a cell viability assay, IC50=1.22579 μM | ||||
| human MEL-HO cell | Growth inhibition assay | Inhibition of human MEL-HO cell growth in a cell viability assay, IC50=1.24238 μM | ||||
| human NKM-1 cell | Growth inhibition assay | Inhibition of human NKM-1 cell growth in a cell viability assay, IC50=1.28636 μM | ||||
| human NCI-H650 cel | Growth inhibition assay | Inhibition of human NCI-H650 cell growth in a cell viability assay, IC50=1.30076 μM | ||||
| human HAL-01 cell | Growth inhibition assay | Inhibition of human HAL-01 cell growth in a cell viability assay, IC50=1.30175 μM | ||||
| human 786-0 cell | Growth inhibition assay | Inhibition of human 786-0 cell growth in a cell viability assay, IC50=1.31283 μM | ||||
| human GI-1 cell | Growth inhibition assay | Inhibition of human GI-1 cell growth in a cell viability assay, IC50=1.31808 μM | ||||
| human Becker cell | Growth inhibition assay | Inhibition of human Becker cell growth in a cell viability assay, IC50=1.36315 μM | ||||
| human CCF-STTG1 cell | Growth inhibition assay | Inhibition of human CCF-STTG1 cell growth in a cell viability assay, IC50=1.42525 μM | ||||
| human MC116 cell | Growth inhibition assay | Inhibition of human MC116 cell growth in a cell viability assay, IC50=1.46122 μM | ||||
| human MDA-MB-468 cell | Growth inhibition assay | Inhibition of human MDA-MB-468 cell growth in a cell viability assay, IC50=1.53736 μM | ||||
| human NB12 cell | Growth inhibition assay | Inhibition of human NB12 cell growth in a cell viability assay, IC50=1.54842 μM | ||||
| human LXF-289 cell | Growth inhibition assay | Inhibition of human LXF-289 cell growth in a cell viability assay, IC50=1.55104 μM | ||||
| human MFE-296 cell | Growth inhibition assay | Inhibition of human MFE-296 cell growth in a cell viability assay, IC50=1.56836 μM | ||||
| human SNU-423 cell | Growth inhibition assay | Inhibition of human SNU-423 cell growth in a cell viability assay, IC50=1.57192 μM | ||||
| human NCI-H2291 cell | Growth inhibition assay | Inhibition of human NCI-H2291 cell growth in a cell viability assay, IC50=1.5761 μM | ||||
| human OCUB-M cell | Growth inhibition assay | Inhibition of human OCUB-M cell growth in a cell viability assay, IC50=1.58314 μM | ||||
| human KU812 cell | Growth inhibition assay | Inhibition of human KU812 cell growth in a cell viability assay, IC50=1.59628 μM | ||||
| human HuO9 cell | Growth inhibition assay | Inhibition of human HuO9 cell growth in a cell viability assay, IC50=1.6128 μM | ||||
| human 639-V cell | Growth inhibition assay | Inhibition of human 639-V cell growth in a cell viability assay, IC50=1.63144 μM | ||||
| human HCC70 cell | Growth inhibition assay | Inhibition of human HCC70 cell growth in a cell viability assay, IC50=1.66146 μM | ||||
| human SNB75 cell | Growth inhibition assay | Inhibition of human SNB75 cell growth in a cell viability assay, IC50=1.66508 μM | ||||
| human D-336MG cell | Growth inhibition assay | Inhibition of human D-336MG cell growth in a cell viability assay, IC50=1.6688 μM | ||||
| human LoVo cell | Growth inhibition assay | Inhibition of human LoVo cell growth in a cell viability assay, IC50=1.73825 μM | ||||
| human EB2 cell | Growth inhibition assay | Inhibition of human EB2 cell growth in a cell viability assay, IC50=1.75019 μM | ||||
| human HSC-2 cell | Growth inhibition assay | Inhibition of human HSC-2 cell growth in a cell viability assay, IC50=1.75727 μM | ||||
| human D-542MG cell | Growth inhibition assay | Inhibition of human D-542MG cell growth in a cell viability assay, IC50=1.85077 μM | ||||
| human COLO-684 cell | Growth inhibition assay | Inhibition of human COLO-684 cell growth in a cell viability assay, IC50=2.20032 μM | ||||
| human GDM-1 cell | Growth inhibition assay | Inhibition of human GDM-1 cell growth in a cell viability assay, IC50=2.24206 μM | ||||
| human SW1088 cell | Growth inhibition assay | Inhibition of human SW1088 cell growth in a cell viability assay, IC50=2.48143 μM | ||||
| human SF295 cell | Growth inhibition assay | Inhibition of human SF295 cell growth in a cell viability assay, IC50=2.76955 μM | ||||
| human NCI-H2030 cell | Growth inhibition assay | Inhibition of human NCI-H2030 cell growth in a cell viability assay, IC50=2.90288 μM | ||||
| human NCI-H1755 cell | Growth inhibition assay | Inhibition of human NCI-H1755 cell growth in a cell viability assay, IC50=3.27063 μM | ||||
| human MDA-MB-361 cell | Growth inhibition assay | Inhibition of human MDA-MB-361 cell growth in a cell viability assay, IC50=3.52709 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 408.45 | Formula | C22H24N4O4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1173900-33-8 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | KIN-193 | Smiles | CC1=CN2C(=O)C=C(N=C2C(=C1)C(C)NC3=CC=CC=C3C(=O)O)N4CCOCC4 | ||
Solubility
|
In vitro |
DMSO : 82 mg/mL ( (200.75 mM) Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Ethanol : 10 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki | |
|---|---|
| In vitro |
AZD6482 also inhibits PI3Kα, γ, and δ, with IC50 of 80 nM to 1.09 μM, which are significantly lower than its (+)-enantiomer (S-form). This compound is an antiplatelet agent; it blocks platelet activation adhesion/aggregation and promotes platelet disaggregation in assay of washed platelet aggregation (WPA), with an IC50 value of 6 nM. Furthermore, by targeting PI3Kβ, this chemical specifically inhibits thrombosis without interfering with normal haemostasis. Therefore, it is used as an anti-thrombotic drug for the prophylaxis of thrombotic disorders. [1]
|
| Kinase Assay |
Assay of PI3K enzyme inhibition
|
|
The inhibition of PI3Kβ, PI3Kα, PI3Kγ, and PI3Kδ is evaluated in an AlphaScreen based enzyme activity assay using human recombinant enzymes. The assay measures PI3K-mediated conversion of PIP2 to PIP3. Biotinylated PIP3, a GST-tagged pleckstrin homology (PH) domain and the two AlphaScreen beads form a complex that elicits a signal upon laser excitation at 680 nm. The PIP3 formed in the enzyme reaction competes with the biotinylated PIP3 for binding to the PH domain thus reducing the signal with increasing enzyme product.
This compound is dissolved in DMSO and added to 384 well plates. PBKβ, PBKα, PBKγ, or PBKδ is added in a Tris buffer (50 mM Tris pH 7.6, 0.05% CHAPS, 5 mM DTT, and 24 mM MgCl2) and allowed to preincubate with this chemical for 20 minutes prior to the addition of substrate solution containing PIP2 and ATP. The enzyme reaction is stopped after 20 minutes by addition of stop solution containing EDTA and biotin-PIP3, followed by addition of detection solution containing GST-grpl PH and AlphaScreen beads. Plates are left for a minimum of 5 hours in the dark prior to analysis. The final concentration of DMSO, ATP and PIP2 in the assay are, 0.8%, 4 μM, and 40 μM, respectively.
IC50 values are calculated according to the equation, y = {a+[(b-a)/(l+(x/IC50)s)]}, where y = % inhibition; a = 0%; b = 100%; s = the slope of the concentration-response curve; x = this compound concentration.
|
References |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































