research use only
PIK-75 HCl PI3K inhibitor
Cat.No.S1205
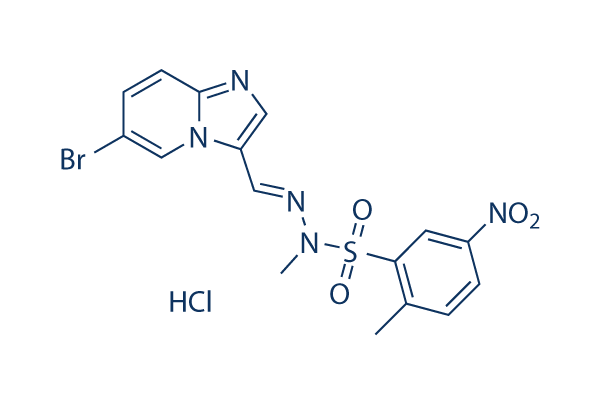
Chemical Structure
Molecular Weight: 488.74
Quality Control
| Related Targets | Akt mTOR GSK-3 ATM/ATR DNA-PK AMPK PDPK1 PTEN PP2A PDK |
|---|---|
| Other PI3K Inhibitors | GDC-0077 (Inavolisib) SAR405 Quercetin (Sophoretin) LY294002 XL147 analogue Tersolisib (STX-478) Buparlisib (BKM120) 740 Y-P (PDGFR 740Y-P) GO-203 TFA Eganelisib (IPI-549) |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human MV4-11 cells | Cytotoxic assay | 72 h | Cytotoxicity against human MV4-11 cells after 72 hrs by CellTiter-Glo assay, IC50=0.003 μM | |||
| human NZOV9 cells | Proliferation assay | Antiproliferative activity against human NZOV9 cells, IC50=0.066 μM | ||||
| human NZB5 cells | Proliferation assay | Antiproliferative activity against human NZB5 cells, IC50=0.069 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 98 mg/mL
(200.51 mM)
Ethanol : 9 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 488.74 | Formula | C16H14BrN5O4S.HCl |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 372196-77-5 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC1=C(C=C(C=C1)[N+](=O)[O-])S(=O)(=O)N(C)N=CC2=CN=C3N2C=C(C=C3)Br.Cl | ||
Mechanism of Action
| Features |
PI3K and DNA-PK inhibitor.
|
|---|---|
| Targets/IC50/Ki |
DNA-PK
(Cell-free assay) 2 nM
p110α
(Cell-free assay) 5.8 nM
p110γ
(Cell-free assay) 76 nM
p110δ
(Cell-free assay) 0.51 μM
|
| In vitro |
PIK-75 shows the impressive potency and isoform selectivity at p110α while the corresponding IC50 values are 1300 nM, 76 nM and 510 nM for other PI3K isoforms, p110β, -γ, and -δ, respectively. Furthermore, when binding to purified p110α, PIK-75 is a noncompetitive inhibitor with respect to ATP with Ki of 36 nM and competitive with respect to the substrate PI with Ki of 2.3 nM. PIK-75 also shows potent inhibition of DNA-PK. PIK-75 (1 μM) reduces cell survival by significantly decreasing mitochondrial activity in unstimulated nonasthmatic airway smooth muscle (ASM) cells, asthmatic ASM cells, and lung fibroblasts. While in TGFβ-stimulated ASM cells, PIK75 only decreases mitochondrial activity in asthmatic cells without effects in nonasthmatic cells. A recent study shows that PIK-75 (10 nM) inhibits TNF-α-induced CD38 mRNA expression and significantly attenuates of TNF-α-induced ADP-ribosyl cyclase activity in human airway smooth muscle cells.
|
| Kinase Assay |
Inhibition Assays
|
|
The PI3K inhibitor PIK-75 is dissolved at 10 mM in dimethyl sulfoxide and stored at −20°C until use. PI3K enzyme activity is determined in 50 μL of 20 mM HEPES, pH 7.5, and 5 mM MgCl2 containing 180 μM phosphatidyl inositol, with the reaction started by the addition of 100 μM ATP (containing 2.5 μCi of [γ-32P]ATP). After a 30-minute incubation at room temperature, the enzyme reaction is stopped by the addition of 50 μL of 1 M HCl. Phospholipids are then extracted with 100 μL of chloroform/methanol [1:1 (v/v)] and 250 μL of 2 M KCl followed by liquid scintillation counting. Inhibitors are diluted in 20% (v/v) dimethyl sulfoxide to generate a concentration versus inhibition of enzyme activity curve, which is then analyzed with the use of Prism version 5.00 for Windows to calculate the IC50. For kinetic analysis, a luminescent assay measuring ATP consumption is used. PI3K enzyme activity is determined in 50 μL of 20 mM HEPES, pH 7.5, and 5 mM MgCl2 with PI and ATP at various concentrations. After a 60-minute incubation at room temperature, the reaction is stopped by the addition of 50 μL of Kinase-Glo followed by a further 15-minute incubation. Luminescence is then read using a Fluostar plate reader. Results are analyzed using Prism.
|
|
| In vivo |
In the ErbB3WT tumor model, PIK-75 reduces in vitro chemotactic response to HRGβ1 and lowers pAkt levels by 40%. Besides, PIK-75 significantly reduces tumor cell motility and in vivo invasion in ErbB3WT primary tumors. In the CD1 male mice, PIK-75 leads to serious impairments in the insulin tolerance test (ITT) and glucose tolerance test (GTT), and an increase in glucose production during a pyruvate tolerance test (PTT).
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
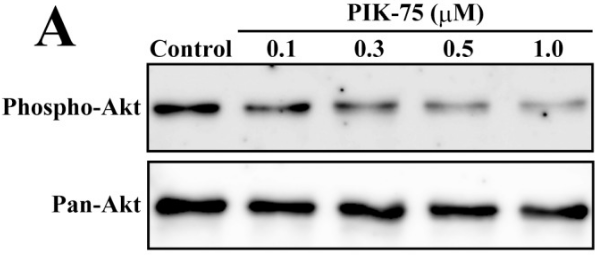
| Western blot | p-AKT / AKT MYCN / p-rpS6 p-AKT / AKT / p-GSK3β / GSK3β / PARP / Survivin / XIAP |
|
23077605 |
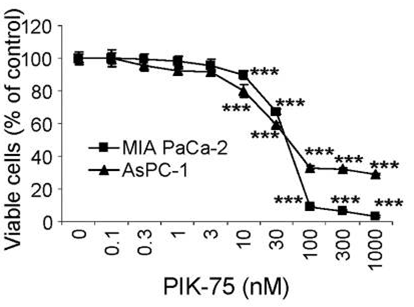
| Growth inhibition assay | Cell viability |
|
24366069 |
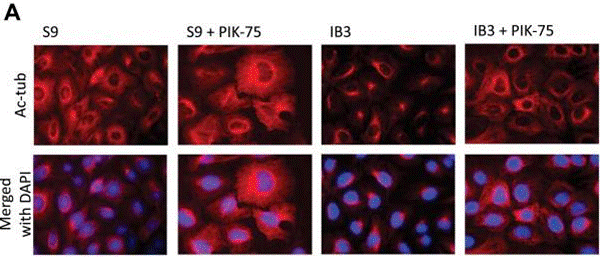
| Immunofluorescence | Ac-tubulin |
|
23873844 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































