- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
Belinostat (PXD101) HDAC inhibitor
Cat.No.S1085
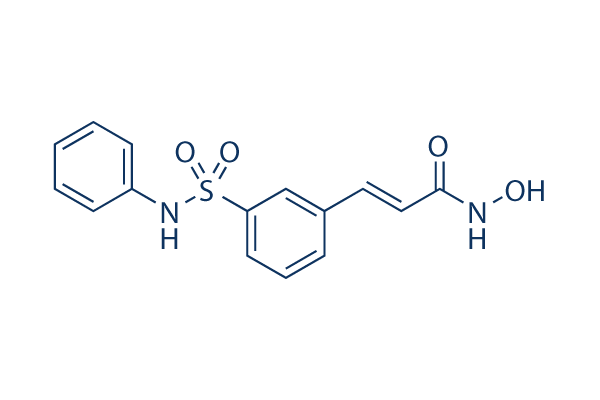
Chemical Structure
Molecular Weight: 318.35
Jump to
Quality Control
Batch:
Purity:
99.74%
99.74
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| HCT116 | Function Assay | 0.9 μM | 24 h | down-regulats TS protein levels after 6 h incubation | 17124594 | |
| HCT116 | Growth Inhibition Assay | 48 h | EC50=0.28 μM | 17124594 | ||
| Granta-519 | Growth Inhibition Assay | 24 h | IC50=56.3 μM | 20068080 | ||
| Jeko-1 | Growth Inhibition Assay | 24 h | IC50=0.2 μM | 20068080 | ||
| HBL-2 | Growth Inhibition Assay | 24 h | IC50=0.4 μM | 20068080 | ||
| Panc-1 | Apoptosis Assay | 100/500/1000 nM | 48 h | induces dose dependent apoptosis | 22681698 | |
| AsPC-1 | Apoptosis Assay | 100/500/1000 nM | 48 h | induces dose dependent apoptosis | 22681698 | |
| T3M4 | Apoptosis Assay | 100/500/1000 nM | 48 h | induces dose dependent apoptosis | 22681698 | |
| Panc-1 | Growth Inhibition Assay | 0-800 nM | 48 h | inhibits cell proliferation in a dose dependent manner | 22681698 | |
| AsPC-1 | Growth Inhibition Assay | 0-800 nM | 48 h | inhibits cell proliferation in a dose dependent manner | 22681698 | |
| T3M4 | Growth Inhibition Assay | 0-800 nM | 48 h | inhibits cell proliferation in a dose dependent manner | 22681698 | |
| MiaPaCa2 | Function Assay | 1/10 μM | 24 h | induces growth arrested in G2/M | 23475695 | |
| AsPc1 | Function Assay | 1/10 μM | 24 h | induces growth arrested in G2/M | 23475695 | |
| Panc0403 | Apoptosis Assay | 1 μM | 24 h | induces apoptosis | 23475695 | |
| Panc1005 | Apoptosis Assay | 1 μM | 24 h | induces apoptosis | 23475695 | |
| Panc0327 | Apoptosis Assay | 1 μM | 24 h | induces apoptosis | 23475695 | |
| Panc0203 | Growth Inhibition Assay | 48 h | EC50=22.2 μM | 23475695 | ||
| PL45 | Growth Inhibition Assay | 48 h | EC50=20.8 μM | 23475695 | ||
| Panc1005 | Growth Inhibition Assay | 48 h | EC50=1.1 μM | 23475695 | ||
| Panc0403 | Growth Inhibition Assay | 48 h | EC50=1.1 μM | 23475695 | ||
| BxPc3 | Growth Inhibition Assay | 48 h | EC50=1.0 μM | 23475695 | ||
| MiaPaCa2 | Growth Inhibition Assay | 48 h | EC50=0.7 μM | 23475695 | ||
| Panc0327 | Growth Inhibition Assay | 48 h | EC50=0.5 μM | 23475695 | ||
| AsPc1 | Growth Inhibition Assay | 48 h | EC50=0.3 μM | 23475695 | ||
| PC9 | Function Assay | 0.5/1/2 μM | 4 h | DMSO | inhibits the levels of Akt (p-Akt) and EGFR | 23515752 |
| H1650 | Function Assay | 0.5/1/2 μM | 4 h | DMSO | inhibits the levels of Akt (p-Akt) and EGFR | 23515752 |
| H460 | Function Assay | 0.5/1/2 μM | 4 h | DMSO | inhibits the levels of Akt (p-Akt) and EGFR | 23515752 |
| PC9 | Function Assay | 500 nM | 24 h | DMSO | decreases EGFR expression | 23515752 |
| H1650 | Function Assay | 500 nM | 24 h | DMSO | decreases EGFR expression | 23515752 |
| H460 | Function Assay | 500 nM | 24 h | DMSO | decreases EGFR expression | 23515752 |
| HCC4006 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.46 μM | 23515752 | |
| HCC2935 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.97 μM | 23515752 | |
| HCC827 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.29 μM | 23515752 | |
| HCC2279 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.4 μM | 23515752 | |
| PC9 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.29 μM | 23515752 | |
| H820 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.4 μM | 23515752 | |
| H1650 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.88 μM | 23515752 | |
| H1975 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.68 μM | 23515752 | |
| H520 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.75 μM | 23515752 | |
| H1299 | Growth Inhibition Assay | 72 h | DMSO | IC50=1.2 μM | 23515752 | |
| H460 | Growth Inhibition Assay | 72 h | DMSO | IC50=0.86 μM | 23515752 | |
| H1666 | Growth Inhibition Assay | 72 h | DMSO | IC50>10 μM | 23515752 | |
| PANC-1 | Function Assay | 10 μM | 2/4 h | DMSO | increases intracellular ROS level | 23743198 |
| PANC-1 | Cell Viability Assay | 1/10 μM | 48 h | DMSO | decreases cell viability in a dose dependent manner | 23743198 |
| PANC-1 | Function Assay | 10 μM | 2/4/6 h | DMSO | induces AMPK activation | 23743198 |
| HL-60 | Function Assay | 0.2 μM | 24/48/72 h | enhances RA-induced granulocytic differentiation | 25864732 | |
| NB4 | Function Assay | 0.2 μM | 24/48/72 h | enhances RA-induced granulocytic differentiation | 25864732 | |
| HL-60 | Function Assay | 2 μM | 24/48 h | blocks cell cycle in S phase | 25864732 | |
| NB4 | Function Assay | 2 μM | 24/48 h | blocks cell cycle in S phase | 25864732 | |
| HL-60 | Cell Viability Assay | 0.2/2 μM | 24/48/72 h | decreases cell viability in both time and dose dependent manner | 25864732 | |
| NB4 | Cell Viability Assay | 0.2/2 μM | 24/48/72 h | decreases cell viability in both time and dose dependent manner | 25864732 | |
| RAW264.7 | Anti-inflammatory assay | 1 hr | Anti-inflammatory activity in LPS-stimulated mouse RAW264.7 cells assessed as suppression of IL6 production pre-incubated for 1 hr before LPS stimulation for 24 hrs by ELISA method, IC50 = 0.000059 μM. | 25113875 | ||
| HEK293 | Function assay | Inhibition of HDAC6 in HEK293 cells, IC50 = 0.015 μM. | 18308563 | |||
| HEK293 | Function assay | Inhibition of HDAC1 in HEK293 cells, IC50 = 0.018 μM. | 18308563 | |||
| HeLa | Function assay | 30 mins | Inhibition of HDAC in human HeLa cells nuclear extracts incubated for 30 mins by fluorescent assay, IC50 = 0.0264 μM. | 25113875 | ||
| HeLa | Function assay | Inhibition of HDAC in human HeLa cells using Fluor de Lys as substrate by fluorescence assay, IC50 = 0.027 μM. | 23639537 | |||
| HeLa | Function assay | Inhibition of HDAC from human HeLa cells, IC50 = 0.028 μM. | 18247554 | |||
| HEK293 | Function assay | Inhibition of HDAC3 in HEK293 cells, IC50 = 0.046 μM. | 18308563 | |||
| MDA-MB-231 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MDA-MB-231 cells after 72 hrs by resazurin dye based fluorescence assay, IC50 = 0.062 μM. | 29456804 | ||
| Jurkat | Antiproliferative assay | 48 hrs | Antiproliferative activity against human Jurkat cells after 48 hrs by MTT assay, IC50 = 0.07 μM. | 29533873 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by resazurin dye based fluorescence assay, IC50 = 0.077 μM. | 29456804 | ||
| HeLa | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HeLa cells after 72 hrs by resazurin dye based fluorescence assay, IC50 = 0.087 μM. | 29456804 | ||
| MCF7 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MCF7 cells after 72 hrs by resazurin dye based fluorescence assay, IC50 = 0.096 μM. | 29456804 | ||
| HEL | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HEL cells after 48 hrs by MTT assay, IC50 = 0.1 μM. | 29533873 | ||
| Huh7 | Antiviral assay | 3 days | Antiviral activity against HCV genotype 1b infected in human Huh7 cells after 3 days by luciferase reporter gene assay, EC50 = 0.12 μM. | 25490700 | ||
| HCT116 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HCT116 cells after 48 hrs by SRB assay, GI50 = 0.13 μM. | 27344487 | ||
| MOLT4 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human MOLT4 cells after 48 hrs by MTT assay, IC50 = 0.14 μM. | 29533873 | ||
| HCT116 | Antiproliferative assay | Antiproliferative activity against human HCT116 cells assessed as growth inhibition, IC50 = 0.16 μM. | 21650221 | |||
| HCT116 | Antiproliferative assay | Antiproliferative activity against human HCT116 cells, IC50 = 0.16 μM. | 21742496 | |||
| SK-N-BE(2) | Antiproliferative assay | 48 hrs | Antiproliferative activity against human SK-N-BE(2) cells after 48 hrs by MTT assay, IC50 = 0.31 μM. | 29533873 | ||
| PC3 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human PC3 cells after 48 hrs by SRB assay, GI50 = 0.39 μM. | 27344487 | ||
| PC3 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human PC3 cells after 96 hrs by celltiter 96 assay, IC50 = 0.45 μM. | 21634430 | ||
| H1299 | Antiproliferative assay | Antiproliferative activity against human H1299 cells, IC50 = 0.46 μM. | 21650221 | |||
| HeLa | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HeLa cells after 48 hrs by MTT assay, IC50 = 0.51 μM. | 29533873 | ||
| HCT116 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human HCT116 cells after 96 hrs by celltiter 96 assay, IC50 = 0.6 μM. | 21634430 | ||
| A2780 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human A2780 cells after 96 hrs by celltiter 96 assay, IC50 = 0.67 μM. | 21634430 | ||
| HuH7 | Cytotoxicity assay | 3 days | Cytotoxicity against human HuH7 cells assessed as inhibition of cell viability after 3 days by CellTiter 96 assay, CC50 = 0.68 μM. | 25490700 | ||
| COLO205 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human COLO205 cells after 96 hrs by celltiter 96 assay, IC50 = 0.7 μM. | 21634430 | ||
| A549 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A549 cells after 48 hrs by SRB assay, GI50 = 0.78 μM. | 27344487 | ||
| HL60 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HL60 cells after 48 hrs by SRB assay, GI50 = 1.09 μM. | 27344487 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50 = 1.1 μM. | 29533873 | ||
| PC3 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human PC3 cells after 48 hrs by MTT assay, IC50 = 1.3 μM. | 29533873 | ||
| NFF | Cytotoxicity assay | 72 hrs | Cytotoxicity against human NFF cells after 72 hrs by SRB assay, IC50 = 1.4 μM. | 28241112 | ||
| HEK293 | Cytotoxicity assay | 48 hrs | Cytotoxicity against HEK293 cells after 48 hrs by resazurin assay, IC50 = 1.4 μM. | 28241112 | ||
| NFF | Cytotoxicity assay | 72 hrs | Cytotoxicity against human NFF cells after 72 hrs by sulforhodamine B assay, IC50 = 1.42 μM. | 30245402 | ||
| HEK293 | Cytotoxicity assay | 48 hrs | Cytotoxicity against HEK293 cells after 48 hrs by resazurin dye based assay, IC50 = 1.42 μM. | 30245402 | ||
| RAW264.7 | Anti-inflammatory assay | 1 hr | Anti-inflammatory activity in LPS-stimulated mouse RAW264.7 cells assessed as suppression of nitric oxide production pre-incubated for 1 hr before LPS stimulation for 24 hrs by Griess reagent based assay, IC50 = 2.2 μM. | 25113875 | ||
| RAW264.7 | Anti-inflammatory assay | 1 hr | Anti-inflammatory activity in LPS-stimulated mouse RAW264.7 cells assessed as suppression of TNFalpha production pre-incubated for 1 hr before LPS stimulation for 24 hrs by ELISA method, IC50 = 4.7 μM. | 25113875 | ||
| RAW264.7 | Anti-inflammatory assay | 1 hr | Anti-inflammatory activity in LPS-stimulated mouse RAW264.7 cells assessed as suppression of PGE2 production pre-incubated for 1 hr before LPS stimulation for 24 hrs by enzyme immunoassay method, IC50 = 8.28 μM. | 25113875 | ||
| Huh-luc/neo7 | Function assay | 1 uM | 1 to 3 hrs | Inhibition of HDAC class 1 in human Huh-luc/neo7 cells assessed as histone H3 acetylation at 1 uM after 1 to 3 hrs by Western blotting analysis | 25937017 | |
| PC3 | Function assay | 0.3 uM | 48 hrs | Inhibition of HDAC in human PC3 cells assessed as increase in amount of acetylated histone H3 at 0.3 uM after 48 hrs by Western blot analysis | 27344487 | |
| HCT116 | Function assay | 0.3 uM | 48 hrs | Inhibition of HDAC in human HCT116 cells assessed as increase in amount of acetylated histone H3 at 0.3 uM after 48 hrs by Western blot analysis | 27344487 | |
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 64 mg/mL
(201.03 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
Dilution Calculator
Molecular Weight Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 318.35 | Formula | C15H14N2O4S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 866323-14-0 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | PXD101,NSC726630, PX-105684 | Smiles | C1=CC=C(C=C1)NS(=O)(=O)C2=CC=CC(=C2)C=CC(=O)NO | ||
Mechanism of Action
| Features |
Lead compound of Topotarget.
|
|---|---|
| Targets/IC50/Ki |
HDAC
(Cell-free assay) 27 nM
|
| In vitro |
Belinostat inhibits the growth of tumor cells (A2780, HCT116, HT29, WIL, CALU-3, MCF7, PC3 and HS852) with IC50 from 0.2-0.66 μM. PD101 shows low activity in A2780/cp70 and 2780AD cells, which are cisplatin and doxorubicin-resistant derivatives of A2780 cells. This compound could induce apoptosis through PARP cleavage and acetylation of histones H3/H4. It inhibits bladder cancer cell growth, especially in 5637 cells, which shows accumulation of G0-G1 phase, decrease in S phase and increase in G2-M phase. The growth inhibitory activity of this chemical on cell lines is not strongly influenced by the multidrug-resistant phenotype, whereas the activity of docetaxel is clearly affected. It could enhance the growth inhibitory activity of docetaxel or carboplatin in OVCAR-3 and A2780 cells. This compound also shows enhanced tubulin acetylation in ovarian cancer cell lines. A recent study shows that it activates protein kinase A in a TGF-β signaling-dependent mechanism and decreases survivin mRNA.
|
| Kinase Assay |
Histone Deacetylase Activity
|
|
Subconfluent cultures are harvested and washed twice in ice cold PBS and pelleted by centrifugation at 200 × g for 5 min. The cell pellet is resuspended in two volumes of lysis buffer [60 mM Tris buffer (pH 7.4) containing 30% glycerol and 450 mM NaCl] and lysed by three freeze (dry ice) thaw (30 °C water bath) cycles. Cell debris is removed by centrifugation at 1.2 × 104 g for 5 min, and the supernatant is stored at −80 °C. Histone H4 peptide (sequence SGRGKGGKGLGKGGAKRHRK corresponding to the 20 NH2-terminal residues) is acetylated by a recombinant protein containing the hypoxanthine-aminopterin-thymidine domain of p300, using [3H]acetyl CoA as a source of acetate. H4 peptide (100 μg) is mixed with hypoxanthine-aminopterin-thymidine buffer (50 mM Tris HCl pH 8.0, 5% glycerol, 50 mM KCl, and 0.1 mM EDTA), 1 mM DTT, 1 mM 4-(2-aminoethyl) benzenesulfonylfluoride, 1 × complete protease inhibitors, 50 μL of purified p300, and 1.85 m [3H]acetyl CoA (4.50Ci/mmol) in a final volume of 300 μL and incubated at 30 °C for 45 min. The p300 protein is removed by incubation with 20 μL of 50% Ni-agaroase beads for 1 hour at 4 °C and centrifugation. The supernatant is applied to a 2 mL Sephadex G15 column, and the flow through is collected. One milliliter of distilled H2O is gently applied, and three drop fractions are collected; this is repeated until 4–5 mL of distilled H2O has been added, and ∼40 fractions are collected. Three microliters of each fraction are diluted in 2 mL of scintillation fluid and counted in a scintillation counter to identify the fractions containing the labeled peptide. These fractions are pooled, and 1 μL of the combined sample is measured to assess the radioactivity in every peptide batch (3-7×103 cpm/μL). For activity assays, the reaction is carried out in a total volume of 150 μL of buffer [60 mM Tris (pH 7.4) containing 30% glycerol] containing 2 μL of cell extract and, where used, 2 μL of this compound. The reaction is started by the addition of 2 μL of [3H] labeled substrate (acetylated histone H4 peptide corresponding to the 20 NH2-terminal residues). Samples are incubated at 37 °C for 45 min, and the reaction stopped by the addition of HCl and acetic acid (0.72 and 0.12 M final concentrations, respectively). Released [3H]acetate is extracted into 750 μL of ethyl acetate, and samples are centrifuged at 1.2× 104 g for 5 min. The upper phase (600 μL) is transferred to 3 mL of scintillation fluid and counted.
|
|
| In vivo |
Belinostat indicates significant tumor growth delay in A2780 and A2780/cp70 xenograft at a dose of 10mg/kg with no effects on the body weight. This compound also induces p21WAF1, HDAC core and cell communication genes in mouse bladder tumors. Its monotherapy induces dose-proportional antitumor effects with TGI of 47% at a dose of 100mg/kg in A2780 xenograft. The combination of this chemical (100 mg/kg) with carboplatin (40 mg/kg) could delay tumor growth from 18.6 days to 22.5 days. Combining with bortezomib, it results in great tumor inhibition and gastrointestinal toxicity in mice with bortezomib-resistant UMSCC-11A xenograft.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-H2AX(Ser139) / KU70 / KU80 / RAD51 / RAD52 / ERCC1 Acetyl Histone H3 / Acetyl Histone H4 / Acetyl tubulin p21 / p27 SOS1 / SOS2 PARP / p-ERK / p-p38 / p38 / p-BRAF / p-MEK / MEK |

|
24155971 |
| Growth inhibition assay | Cell viability IC50 |
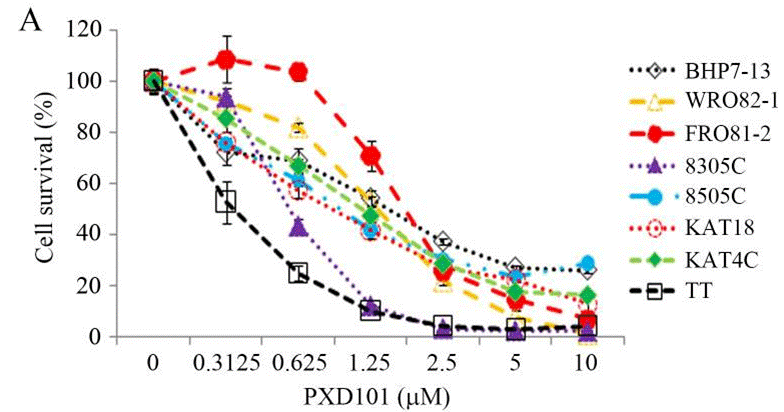
|
24155971 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT06406465 | Not yet recruiting | Carcinoma Neuroendocrine|Tumor Neuroendocrine|Tumors Neuroendocrine|Neuroendocrine; Carcinoma|Small Cell; Receptors |
National Cancer Institute (NCI)|National Institutes of Health Clinical Center (CC) |
May 15 2024 | Phase 2 |
| NCT04315233 | Recruiting | Metastatic Breast Cancer|Recurrent Ovarian Carcinoma |
University of Utah|Novartis|Acrotech Biopharma |
May 3 2021 | Phase 1 |
| NCT04703920 | Recruiting | Metastatic Breast Cancer|Metastatic Castration-resistant Prostate Cancer|Metastatic Ovarian Carcinoma |
University of Michigan Rogel Cancer Center|Pfizer|Acrotech Biopharma Inc. |
March 4 2021 | Phase 1 |
| NCT03772925 | Active not recruiting | Recurrent Acute Myeloid Leukemia|Recurrent Myelodysplastic Syndrome|Refractory Acute Myeloid Leukemia|Refractory Myelodysplastic Syndrome |
National Cancer Institute (NCI) |
June 20 2019 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
Could you please give some suggestions for the use of it in vivo (i.p. injection)?
Answer:
For I.P. injection, it can be dissolved in 2% DMSO+30% PEG 300+ddH2O at 10 mg/ml clearly. When preparing the solution, please dissolve this compound in DMSO clearly first. Then add PEG, after they mixed well, then dilute with water. Hope this information is useful to you.






































