research use only
Dinaciclib (SCH 727965) CDK inhibitor
Cat.No.S2768
-chemical-structure-s2768.gif)
Chemical Structure
Molecular Weight: 396.49
Quality Control
Products Often Used Together with Dinaciclib (SCH 727965)
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| CA46 | Apoptosis Assay | 100 nM | 24 h | induces cell cycle arrest | 25289887 | |
| Kasumi-1 | Apoptosis Assay | 100 nM | 24 h | induces cell cycle arrest | 25289887 | |
| U937 | Function Assay | 2/5/10 nM | 3 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| 8226 | Function Assay | 2/5/10 nM | 4 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| H929 | Function Assay | 2/5/10 nM | 4 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| K562 | Function Assay | 1.5/3/8 nM | 6 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| BaF3/Bcr-abl | Function Assay | 1.5/3/8 nM | 6 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| U937 | Function Assay | 2/10 nM | 3 h | blocks induction of XBP-1s and downstream targets | 24362465 | |
| 1205Lu | Growth Inhibition Assay | 10/30 nM | 72 h | inhibits cell growth and survival | 23527225 | |
| WM1366 | Growth Inhibition Assay | 10/30 nM | 72 h | inhibits cell growth and survival | 23527225 | |
| RD | Growth Inhibition Assay | IC50=8.2 nM | 22315240 | |||
| Rh41 | Growth Inhibition Assay | IC50=10.5 nM | 22315240 | |||
| Rh18 | Growth Inhibition Assay | IC50=10.5 nM | 22315240 | |||
| Rh30 | Growth Inhibition Assay | IC50=9 nM | 22315240 | |||
| BT-12 | Growth Inhibition Assay | IC50=8.5 nM | 22315240 | |||
| CHLA-266 | Growth Inhibition Assay | IC50=7.3 nM | 22315240 | |||
| TC-71 | Growth Inhibition Assay | IC50=3.9 nM | 22315240 | |||
| CHLA-9 | Growth Inhibition Assay | IC50=8 nM | 22315240 | |||
| CHLA-10 | Growth Inhibition Assay | IC50=6.3 nM | 22315240 | |||
| CHLA-258 | Growth Inhibition Assay | IC50=9.9 nM | 22315240 | |||
| GBM2 | Growth Inhibition Assay | IC50=6.5 nM | 22315240 | |||
| NB-1643 | Growth Inhibition Assay | IC50=3.3 nM | 22315240 | |||
| NB-EBc1 | Growth Inhibition Assay | IC50=7 nM | 22315240 | |||
| CHLA-90 | Growth Inhibition Assay | IC50=7.5 nM | 22315240 | |||
| CHLA-136 | Growth Inhibition Assay | IC50=9.8 nM | 22315240 | |||
| NALM-6 | Growth Inhibition Assay | IC50=4.6 nM | 22315240 | |||
| COG-LL-317 | Growth Inhibition Assay | IC50=6.5 nM | 22315240 | |||
| RS4;11 | Growth Inhibition Assay | IC50=5.1 nM | 22315240 | |||
| MOLT-4 | Growth Inhibition Assay | IC50=9.3 nM | 22315240 | |||
| CCRF-CEM | Growth Inhibition Assay | IC50=5.6 nM | 22315240 | |||
| Kasumi-1 | Growth Inhibition Assay | IC50=4.5 nM | 22315240 | |||
| Karpas-299 | Growth Inhibition Assay | IC50=3.9 nM | 22315240 | |||
| Ramos-RA1 | Growth Inhibition Assay | IC50=7.9 nM | 22315240 | |||
| MIAPaCa-2 | Growth Inhibition Assay | 72 h | GI50=10 nM | 21768779 | ||
| Pa20C | Growth Inhibition Assay | 72 h | GI50=20 nM | 21768779 | ||
| ML-1 | Apoptosis Assay | 1-1000 nM | 4 h | induces apoptosis slightly | 21768777 | |
| Cytotoxicity assay | MDA-MB-436 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | NCI-H929 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | MDA-MB-231 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | SK-ES-1 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | A673 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | SK-BR-3 | 72 hrs | IC50 = 0.005 μM | 23600925 | ||
| Cytotoxicity assay | MNNG-HOS | 72 hrs | IC50 = 0.0055 μM | 23600925 | ||
| Cytotoxicity assay | SK-UT-1 | 72 hrs | IC50 = 0.006 μM | 23600925 | ||
| Cytotoxicity assay | U266 | 72 hrs | IC50 = 0.006 μM | 23600925 | ||
| Cytotoxicity assay | RPMI18226 | 72 hrs | IC50 = 0.009 μM | 23600925 | ||
| Cytotoxicity assay | SW872 | 72 hrs | IC50 = 0.0095 μM | 23600925 | ||
| Cytotoxicity assay | T47D | 72 hrs | IC50 = 0.01 μM | 23600925 | ||
| Apoptosis assay | A673 | 24 hrs | EC50 = 0.011 μM | 23600925 | ||
| Cytotoxicity assay | MCF7 | 72 hrs | IC50 = 0.02 μM | 23600925 | ||
| Function assay | Sf9 | IC50 = 0.072 μM | 26741853 | |||
| Function assay | sf9 | IC50 = 0.002 μM | 26851505 | |||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.003 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.003 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.004 μM | 27171036 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.004 μM | 27171036 | ||
| Function assay | Sf9 | 10 uM | IC50 = 0.004 μM | 29329658 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 29853338 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.001 μM | 29853338 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.003 μM | 29853338 | ||
| Function assay | Sf9 | 1 hr | IC50 = 0.004 μM | 29853338 | ||
| Antiproliferative assay | MOLM13 | 72 hrs | GI50 = 0.0033 μM | 30253346 | ||
| Antiproliferative assay | MEC1 | 72 hrs | GI50 = 0.0036 μM | 30253346 | ||
| Antiproliferative assay | MOLM14 | 72 hrs | GI50 = 0.0045 μM | 30253346 | ||
| Antiproliferative assay | COLO205 | 72 hrs | GI50 = 0.0068 μM | 30253346 | ||
| Antiproliferative assay | HL60 | 72 hrs | GI50 = 0.008 μM | 30253346 | ||
| Antiproliferative assay | Ramos | 72 hrs | GI50 = 0.0086 μM | 30253346 | ||
| Antiproliferative assay | GISTT1 | 72 hrs | GI50 = 0.0088 μM | 30253346 | ||
| Antiproliferative assay | U937 | 72 hrs | GI50 = 0.01 μM | 30253346 | ||
| Antiproliferative assay | A431 | 72 hrs | GI50 = 0.011 μM | 30253346 | ||
| Antiproliferative assay | SKM1 | 72 hrs | GI50 = 0.011 μM | 30253346 | ||
| Antiproliferative assay | MEC2 | 72 hrs | GI50 = 0.011 μM | 30253346 | ||
| Antiproliferative assay | A375 | 72 hrs | GI50 = 0.011 μM | 30253346 | ||
| Antiproliferative assay | OCI-AML3 | 72 hrs | GI50 = 0.013 μM | 30253346 | ||
| Antiproliferative assay | BE(2)-M17 | 72 hrs | GI50 = 0.021 μM | 30253346 | ||
| Antiproliferative assay | CHO | 72 hrs | GI50 = 0.16 μM | 30253346 | ||
| Function assay | NCI-H929 | 0.005 uM | 24 hrs | Inhibition of CDK2-mediated Rb phosphorylation at Ser 807/811 in human NCI-H929 cells at 0.005 uM after 24 hrs by immunoblotting analysis | 23600925 | |
| Function assay | A673 | 0.05 uM | 24 hrs | Inhibition of CDK2-mediated Rb phosphorylation at Ser 807/811 in human A673 cells at 0.05 uM after 24 hrs by immunoblotting analysis | 23600925 | |
| Apoptosis assay | MEC1 | 0.01 uM | 24 hrs | Induction of apoptosis in human MEC1 cells assessed as decrease in MCL-1 level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Apoptosis assay | HL60 | 0.01 uM | 24 hrs | Induction of apoptosis in human HL60 cells assessed as decrease in MCL-1 level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Apoptosis assay | MV4-11 | 0.01 uM | 24 hrs | Induction of apoptosis in human MV4-11 cells assessed as decrease in c-MYC level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Apoptosis assay | MEC1 | 0.01 uM | 24 hrs | Induction of apoptosis in human MEC1 cells assessed as decrease in c-MYC level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Apoptosis assay | MV4-11 | 0.01 uM | 24 hrs | Induction of apoptosis in human MV4-11 cells assessed as decrease in MCL-1 level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Apoptosis assay | HL60 | 0.01 uM | 24 hrs | Induction of apoptosis in human HL60 cells assessed as decrease in c-MYC level at 0.01 uM after 24 hrs by immunoblotting analysis | 30253346 | |
| Cytotoxicity assay | U2OS | 96 hrs | IC50 = 0.006 μM | ChEMBL | ||
| Function assay | U2OS | 1 hr | IC50 = 0.007 μM | ChEMBL | ||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 79 mg/mL
(199.24 mM)
Ethanol : 35 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 396.49 | Formula | C21H28N6O2 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 779353-01-4 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | SCH727965, PS-095760 | Smiles | CCC1=C2N=C(C=C(N2N=C1)NCC3=C[N+](=CC=C3)[O-])N4CCCCC4CCO | ||
Mechanism of Action
| Targets/IC50/Ki |
CDK2
(Cell-free assay) 1 nM
CDK5
(Cell-free assay) 1 nM
CDK1
(Cell-free assay) 3 nM
CDK9
(Cell-free assay) 4 nM
|
|---|---|
| In vitro |
Dinaciclib is also a potent DNA replication inhibitor that blocks thymidine (dThd) DNA incorporation in A2780 cells with IC50 of 4 nM. This compound strongly suppresses phosphorylation of Rb on Ser 807/811 at concentrations >6.25 nM, which is in agreement with the observation that 4 nM concentrations are required for 50% inhibition of dThd DNA incorporation in the same cell model. Significantly, complete suppression of Rb phosphorylation is correlated with the onset of apoptosis, as indicated by the appearance of the p85 PARP cleavage product in cells exposed to >6.25 nM of this chemical. It is active against a broad spectrum of human tumor cell lines. Addition of this compound during exposure also suppresses accumulation of γ-H2AX, in a dose-dependent manner. It inhibits melanoma cell proliferation, and drives melanoma cells into massive apoptosis. This chemical induces the apoptosis of several osteosarcoma cell lines including those resistant to doxorubicin. It attenuates the phosphorylation of RNAP II at serine 2 and the phosphorylation of the CDK inhibitor p27Kip1 at threonine 187. Reductions in phosphorylation activity occurrs at 12 - 40 nM of this compound (4 to 16 hours post-addition). It also reduces the phosphorylation of Rb at serine 807/811. This chemical induces the apoptosis of mock- and p53-depleted U2OS cells to a similar extent. |
| Kinase Assay |
Cyclin/CDK kinase assay
|
|
Recombinant cyclin/CDK holoenzymes are purified from Sf9 cells engineered to produce baculoviruses that express a specific cyclin or CDK. Cyclin/CDK complexes are typically diluted to a final concentration of 50 μg/mL in a kinase reaction buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, and 0.1 mM sodium orthovanadate. For each kinase reaction, 1 μg of enzyme and 20 μL of a 2-μM substrate solution (a biotinylated peptide derived from histone H1) are mixed and combined with 10 μL of diluted this compound. The reaction is started by the addition of 50 μL of 2 μM ATP and 0.1 μCi of 33P-ATP. Kinase reactions are incubated for 1 hour at room temperature and are stopped by the addition of 0.1% Triton X-100, 1 mM ATP, 5 mM EDTA, and 5 mg/mL streptavidin-coated SPA beads. SPA beads are captured using a 96-well GF/B filter plate and a Filtermate universal harvester. Beads are washed twice with 2 M NaCl and twice with 2 M NaCl containing 1% phosphoric acid. The signal is then assayed using a TopCount 96-well liquid scintillation counter.
|
|
| In vivo |
Dinaciclib i.p. administration at 8, 16, 32, and 48 mg/kg daily for 10 days results in tumor inhibition by 70%, 70%, 89%, and 96%, respectively. This compound's MED (minimum effective dose) appears to be <8 mg/kg. It is well tolerated, and the maximum body weight loss in the highest dosage group is 5%. This chemical has dose-dependent antitumor activity in vivo, and that nearly complete inhibition of tumor growth occurs at a dose level below the MTD (maximum tolerated dose). It has a short plasma half-life in mouse. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | Cleaved PARP / c-Myc Mcl-1 / Bcl-2 / Bcl-xl / Bax / Bak / PUMA / Noxa RNAP II (P-Ser2/P-Ser5) Survivin |
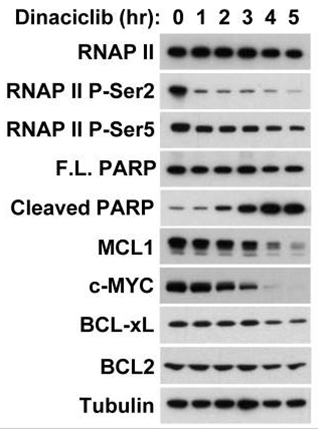
|
25289887 |
| Growth inhibition assay | Cell viability Cell viability |
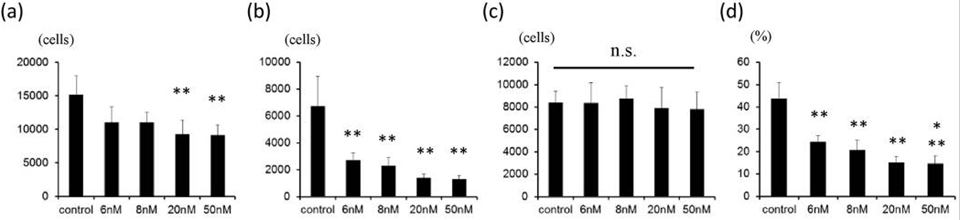
|
28361959 |
| Immunofluorescence | cyclin B1 / α-tubulin / Aurora A OCT4 |
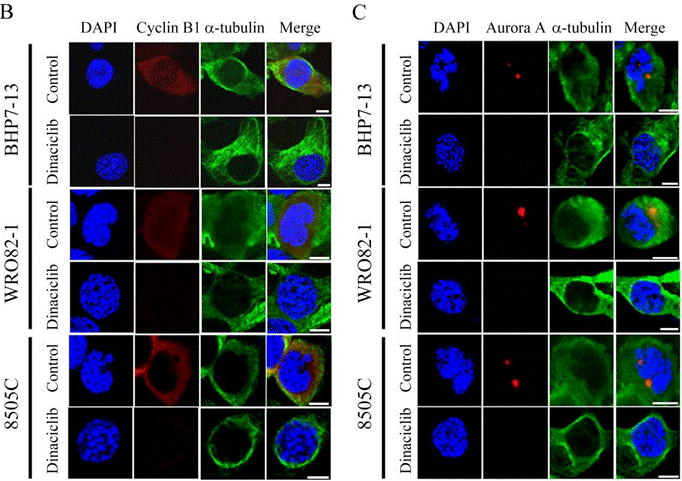
|
28207834 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT03484520 | Terminated | Cancer - Acute Myeloid Leukemia |
AbbVie|Merck Sharp & Dohme LLC |
July 23 2018 | Phase 1 |
| NCT01434316 | Active not recruiting | Advanced Malignant Solid Neoplasm |
National Cancer Institute (NCI) |
November 1 2011 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
I want to know how to reconstitute it for in vivo studies?
Answer:
It can be dissolved in 2% DMSO/30% PEG 300/ddH2O at 10 mg/ml as a clear solution for injection. And this compound in 15% Captisol at 8 mg/ml is a suspension for oral administration.






































