research use only
Fidaxomicin DNA/RNA Synthesis inhibitor
Cat.No.S4227
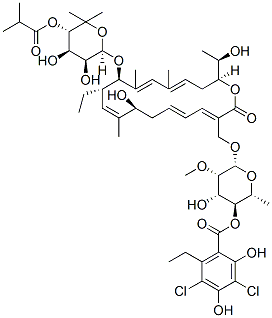
Chemical Structure
Molecular Weight: 1058.04
Quality Control
| Related Targets | HDAC PARP ATM/ATR DNA-PK WRN Topoisomerase PPAR Sirtuin Casein Kinase eIF |
|---|---|
| Other DNA/RNA Synthesis Inhibitors | CX-5461 (Pidnarulex) B02 SCR7 Favipiravir (T-705) EED226 BMH-21 RK-33 Triapine (3-AP) Carmofur YK-4-279 |
Solubility
|
In vitro |
DMSO
: 100 mg/mL
(94.51 mM)
Ethanol : 6 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 1058.04 | Formula | C52H74Cl2O18 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 873857-62-6 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | OPT-80, PAR-101 | Smiles | CCC1C=C(C(CC=CC=C(C(=O)OC(CC=C(C=C(C1OC2C(C(C(C(O2)(C)C)OC(=O)C(C)C)O)O)C)C)C(C)O)COC3C(C(C(C(O3)C)OC(=O)C4=C(C(=C(C(=C4O)Cl)O)Cl)CC)O)OC)O)C | ||
Mechanism of Action
| Targets/IC50/Ki |
RNA polymerase
|
|---|---|
| In vitro |
Fidaxomicin acts as a RNA polymerase inhibitor by binding to the DNA template–RNA polymerase (RNAP) complex prior to the formation of the open RNAP-DNA complex in which transcription is initiated. Therefore this compound will inhibit protein synthesis. As a result, apoptosis is triggered in susceptible organisms such as C. difficile. |
| In vivo |
The minimum inhibitory concentration for 90% of organisms for fidaxomicin against C. difficile is 0.9978 to 2 μg/mL. This compound is not systemically absorbed as shown by a plasma concentrations below the lower limit of quantification after single-dose or multiple-dose. In contrast, fecal concentrations of this chemical are much higher and are concentration-dependent. Cmax = 2 hours; Tmax = 5.2 ng/mL; AUC = 14 ng•hr/mL. It is hydrolyzed by gastric acid or intestinal microsomes into a less active metabolite (OP-1118). The cytochrome enzyme system are not involved in the metabolism of this compound. |
References |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT04138706 | Recruiting | Clostridium Difficile Infection |
McGill University Health Centre/Research Institute of the McGill University Health Centre|Canadian Institutes of Health Research (CIHR) |
November 19 2020 | Phase 3 |
| NCT02437591 | Completed | Inflammatory Bowel Disease (IBD)|Clostridium Difficile Infection (CDI) |
Astellas Pharma Europe Ltd.|Astellas Pharma Inc |
August 13 2015 | Phase 4 |
| NCT02395848 | Terminated | Clostridium Difficile Infection |
McMaster University |
July 2015 | Phase 3 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































