research use only
Selisistat (EX-527) SIRT1 inhibitor
Cat.No.S1541
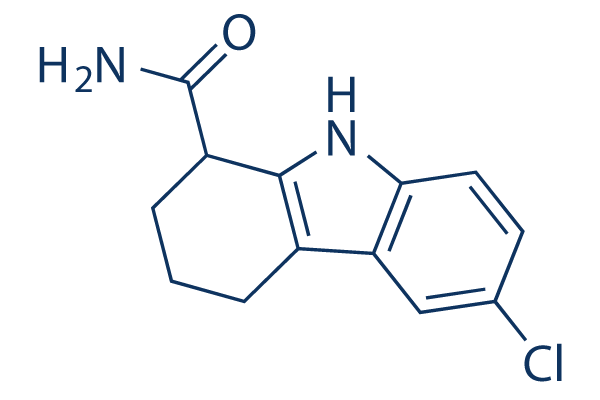
Chemical Structure
Molecular Weight: 248.71
Quality Control
| Related Targets | HDAC JAK BET Histone Methyltransferase PKC PARP HIF PRMT EZH2 AMPK |
|---|---|
| Other Sirtuin Inhibitors | SRT 1720 Hydrochloride Sirtinol Fisetin 3-TYP AGK2 SRT2104 (GSK2245840) OSS_128167 SirReal2 Thiomyristoyl NRD167 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| Platelets | Apoptosis Assay | 10/50 μM | 10 min | DMSO | increases ROS level in a dose-dependent manner | 25829495 |
| HEK 293 | Growth Inhibition Assay | 24 h | IC50=97.7 ± 8.1 μM | 24998427 | ||
| HeLa | Growth Inhibition Assay | 24 h | IC50=37.9 ± 1.8 μM | 24998427 | ||
| HEK 293 | Growth Inhibition Assay | 48 h | IC50=69.0 ± 0.7 μM | 24998427 | ||
| HeLa | Growth Inhibition Assay | 48 h | IC50=8.9 ± 1.9 μM | 24998427 | ||
| RMECs | Apoptosis Assay | 10 μM | 24 h | attenuates the anti-apoptotic effect of Des-G | 24486147 | |
| HUVECs | Apoptosis Assay | 10 μM | 24 h | bolishes the protective effect of resveratrol in cell viability | 23358928 | |
| K562 | Function Assay | 0.1-1 μM | 2 h | stimulates Nrf2-dependent gene transcription | 21196497 | |
| INS-1E | Function Assay | 1 μm | 24 h | prevents resveratrol-induced up-regulation of Glut2, glucokinase,Pdx-1, and Tfam | 21163946 | |
| MCF-7 | Growth Inhibition Assay | 0-100 μM | 24/48/72 h | DMSO | represses cell proliferation at the concentration ≥100 μM | 20371709 |
| NCI-H460 | Function Assay | 1 μm | 6 h | DMSO | produces a concentration-dependent increase in the amount of acetylated p53 | 16354677 |
| 293T | Function assay | Inhibition of human recombinant GST-tagged SIRT1 expressed in 293T cells by Fluor de Lys fluorescence assay, IC50 = 0.038 μM. | 21306906 | |||
| Escherichia coli cells | Function assay | Inhibition of human recombinant SIRT1 expressed in Escherichia coli cells using acetylated Lys side chain amino acids 379-382 (Arg-His-Lys-Lys(Ac)) p53 conjugated with aminomethylcoumarin as substrate by fluorescence assay, IC50 = 0.16 μM. | 22931526 | |||
| BL21 (DE3) | Function assay | Inhibition of full length human SIRT1 expressed in Escherichia coli BL21 (DE3) cells using fluorogenic 7-amino-4-methylcoumarin (AMC)-labeled peptide by fluorescence assay, IC50 = 0.21 μM. | 25275824 | |||
| Namalwa cells | Function assay | 3 hrs | Inhibition of SIRT1-mediated endogenous p53 deacetylase activity in human Namalwa cells assessed as concentration required to enhance p53 deacetylation to twice the basal level after 3 hrs by ELISA, INH = 0.6 μM. | 25971769 | ||
| BL21 (DE3) | Function assay | Inhibition of full length human SIRT2 expressed in Escherichia coli BL21 (DE3) cells using fluorogenic 7-amino-4-methylcoumarin (AMC)-labeled peptide by fluorescence assay, IC50 = 1.94 μM. | 25275824 | |||
| Escherichia coli cells | Function assay | Inhibition of human recombinant SIRT2 expressed in Escherichia coli cells using acetylated Lys side chain amino acids 379-382 (Arg-His-Lys-Lys(Ac)) p53 conjugated with aminomethylcoumarin as substrate by fluorescence assay, IC50 = 48.5 μM. | 22931526 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| Hs683 | Cell cycle assay | 24 to 48 hrs | Cell cycle arrest in human Hs683 cells assessed as accumulation at G1 phase at IC50 after 24 to 48 hrs by propidium iodide staining-based flow cytometry | 28475330 | ||
| HCT116 | Function assay | 10 uM | 8 hrs | Inhibition of SIRT1 in human HCT116 cells assessed as increase in acetylated p53 at 10 uM after 8 hrs by Western blot analysis | 22642300 | |
| NCI-H460 | Function assay | 6 hrs | Inhibition of SIRT-2 in human NCI-H460 cells assessed as inhibition of p53 deacetylation after 6 hrs by immunoprecipitation/immunoblotting analysis, IC50 = 1 μM. | ChEMBL | ||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 300 mg/mL
(1206.22 mM)
Ethanol : 30 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 248.71 | Formula | C13H13ClN2O |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 49843-98-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | SEN0014196 | Smiles | C1CC(C2=C(C1)C3=C(N2)C=CC(=C3)Cl)C(=O)N | ||
Mechanism of Action
| Features |
Greater potency, specificity, stability, and lower toxicity than other inhibitors of SIRT1 catalytic activity identified to date.
|
|---|---|
| Targets/IC50/Ki |
SIRT1
(Cell-free assay) 38 nM
|
| In vitro |
Selisistat (EX 527) exhibits a potently inhibitory effect against SIRT1 deacetylase activity in a concentration-dependent manner with an IC50 of 38 nM, and displays much lower activity against SIRT2 and SIRT3 with IC50 values of 19.6 μM and 48.7 μM, respectively. It does not inhibit SIRT4-7 and class I/II HDAC activity at concentrations up to 100 μM. This compound alone (1 μM) has no detectable effect on the acetylation of p53 lysine 382 in NCI-H460 cells, but significantly increases the amount of acetylated p53 in NCI-H460 cells, human mammary epithelial cells, U-2 OS and MCF-7 cells subjected to genotoxic agents Hydrogen peroxide, which is more effective than that caused by Nicotinamide (5 mM). Surprisingly, it does not result in detectable effects on p53-controled gene expression, cell survival, or cell proliferation. It causes a 90% increase in cell number of HCT116 cells after 7 days in the condition of 0.1% serum but not 10% serum, suggesting that SIRT1 is a significant regulator of cell proliferation during growth factor deprivation conditions. Additionally, it abrogates resveratrol effects on glucose responses, and prevents resveratrol-induced up-regulation of Glut2, glucokinase, Pdx-1, and Tfam in INS-1E Cells, due to the opposite effect of EX 527 and resveratrol on SIRT1 deacetylase activity. |
| Kinase Assay |
Inhibition of GST-SIRT1 deacetylase activity
|
|
293T cells are transiently transfected with GST-tagged human SIRT1 in the pDEST27 Gateway vector using FuGENE-6. After 48 hours, the cells are lysed with 50 mM Tris, pH 8.0, 120 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40, supplemented with Complete Mini protease inhibitor cocktail tablets. GST-SIRT1 is purified from lysates and washed extensively in the above buffer. The deacetylation assay is performed with approximately 30 ng of GST-SIRT1 in the presence of Selisistat (EX 527) (48 pM to 100 μM). Deacetylation is measured using the Fluor de Lys kit using a fluorogenic peptide encompassing residues 379 to 382 of p53, acetylated on lysine 382. The acetylated lysine residue is coupled to an aminomethylcoumarin moiety. The peptide is deacetylated by SIRT1, followed by the addition of a proteolytic developer that releases the fluorescent aminomethylcoumarin. Briefly, enzyme preparations are incubated with 170 μM NAD+ and 100 μM p53 fluorogenic peptide for 45 minutes at 37 °C followed by incubation in developer for 15 minutes at 37 °C. Fluorescence is measured by excitation at 360 nm and emission at 460 nm and enzymatic activity is expressed in relative fluorescence units.
|
|
| In vivo |
Administration of Selisistat (EX 527; ~10 μg) to rats increases hypothalamic acetyl-p53 levels by inhibiting hypothalamic SIRT1 activity. Co-administration of this compound with ghrelin markedly blunts the orexigenic action of ghrelin by decreasing the pAMPK levels, increasing the ACC levels, and abolishing the higher expression of the transcription factors FoxO1, pCREB, and Bsx and the neuropeptides NPY and AgRP in the hypothalamic arcuate nucleus. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
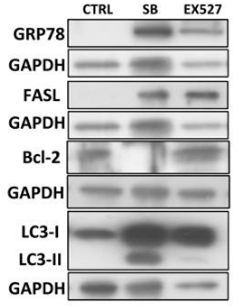
| Western blot | GRP78 / FASL / Bcl-2 / LC3 Ac-H3K9 / Fibronectin / Collagen 1 / α-SMA PCNA / Cyclin D1 / Cyclin E pEGFR / EGFR / pPDGFRβ / PDGFRβ pSTAT3 / STAT3 p27 FOXO3a / SIRT1 / Ac-p53 / p16(INK4a) |
|
29312612 |
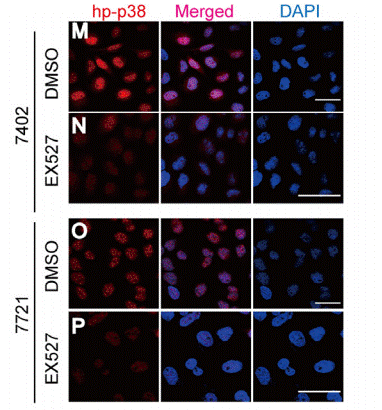
| Immunofluorescence | p-p38 |
|
26824501 |
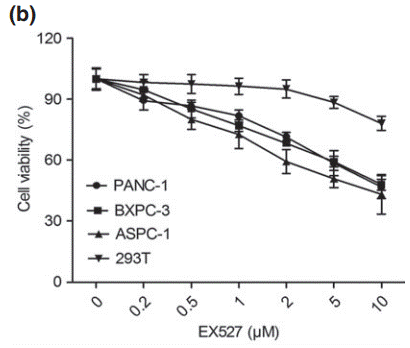
| Growth inhibition assay | Cell viability |
|
24484175 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT01485965 | Completed | Huntington''s Disease |
Siena Biotech S.p.A. |
November 2011 | Phase 1 |
| NCT01521832 | Completed | Huntington''s Disease |
Siena Biotech S.p.A. |
October 2009 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
what is the extinction coefficient of it?
Answer:
Its extinction coefficient is 1421.650635.






































