research use only
MM-102 (HMTase Inhibitor IX) WDR5/MLL1 Inhibitor
Cat.No.S7265
Chemical Structure
Molecular Weight: 669.8
Quality Control
| Related Targets | HDAC JAK BET PKC PARP HIF PRMT EZH2 AMPK Histone Acetyltransferase |
|---|---|
| Other Histone Methyltransferase Inhibitors | Pinometostat (EPZ5676) 3-Deazaneplanocin A (DZNep) Hydrochloride BIX-01294 Trihydrochloride UNC1999 EPZ015666 (GSK3235025) EPZ004777 Chaetocin SGC 0946 EPZ005687 UNC0638 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| Rosetta 2 (DE3) pLysS | Function assay | Inhibition of MLL1 binding to N-terminal His-tagged WRD5 23 deletion mutant (24 to 334 residues) (unknown origin) expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells by fluorescence polarization assay, Ki = 0.001 μM. | 29254892 | |||
| Rosetta 2 (DE3) pLysS | Function assay | Inhibition of MLL1 binding to N-terminal His-tagged WRD5 23 deletion mutant (24 to 334 residues) (unknown origin) expressed in Escherichia coli Rosetta 2 (DE3) pLysS cells by fluorescence polarization assay, IC50 = 0.0024 μM. | 29254892 | |||
| Rosetta2-(DE3) pLysS | Function assay | 5 hrs | Inhibition of C-terminal 5-FAM-WIN peptide binding to N-terminal His-tagged WDR5 (24 to 334 residues) (unknown origin) expressed in Rosetta2-(DE3) pLysS cells after 5 hrs by fluorescence polarization assay, IC50 = 0.0024 μM. | 27720555 | ||
| MV4-11 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MV4-11 cells harboring MLL-AF4 assessed as inhibition of cell viability after 72 hrs by CCK8-based colorimetric assay, IC50 = 20.7 μM. | 27720555 | ||
| MV4-11 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MV4-11 cells harboring MLL-AF4 fusion protein after 72 hrs by CCK8 assay, IC50 = 20.7 μM. | 27598236 | ||
| MV4-11 | Antiproliferative assay | 7 days | Antiproliferative activity against human MV4-11 cells harboring MLL-AF4 after 7 days by CellTiter-Glo luminescent cell viability Assay, IC50 = 25 μM. | 27720555 | ||
| MV4-11 | Antiproliferative assay | Antiproliferative activity against human MV4-11 cells harboring MLL-AF4 fusion protein by CellTiter-Glo assay, IC50 = 25 μM. | 27598236 | |||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells harboring BCR-ABL assessed as inhibition of cell viability after 72 hrs by CCK8-based colorimetric assay, IC50 = 37.8 μM. | 27720555 | ||
| K562 | Growth inhibition assay | 7 days | Growth inhibition of human K562 cells after 7 days by MTS assay, IC50 = 37.8 μM. | 27598236 | ||
| MV4-11 | Function assay | 10 uM | 7 days | Inhibition of MLL1-WDR5 protein-protein interaction in human MV4-11 cells harboring MLL-AF4 fusion protein assessed as inhibition of MLL1 histone methyltransferase activity by measuring reduction in H3K4me2 level at 10 uM after 7 days by Western blot meth | 27598236 | |
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 100 mg/mL
(149.29 mM)
Ethanol : 100 mg/mL Water : 50 mg/mL |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 669.8 | Formula | C35H49F2N7O4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1417329-24-8 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | HMTase Inhibitor IX | Smiles | CCC(CC)(C(=O)NC(CCCN=C(N)N)C(=O)NC1(CCCC1)C(=O)NC(C2=CC=C(C=C2)F)C3=CC=C(C=C3)F)NC(=O)C(C)C | ||
Mechanism of Action
| Targets/IC50/Ki |
WDR5
(Cell-free assay) 2.4 nM
|
|---|---|
| In vitro |
MM-102, as a MLL1 mimetic, shows high binding affinities to WDR5 with IC50 of 2.9 nM and Ki of < 1 nM. In the MLL1-AF9 transduced murine cells, this compound specifically reduces expression of two critical MLL1 target genes (HoxA9 and Meis-1), which are required for MLL1 mediated leukemogenesis. In addition, it effectively and selectively inhibits cell growth and induces apoptosis in leukemia cells harboring MLL1 fusion proteins.
|
| Kinase Assay |
In Vitro Histone Methyltransferase (HMT) Assay
|
|
The HMT assay is performed in 50 mM HEPES pH 7.8, 100 mM NaCl, 1.0 mM EDTA, and 5% glycerol at 22 °C. Each reaction contains 1.5 μCi of the co-factor, 3H-S-adenosylmethionine. H3 10-residue peptide is used as the substrate at 50 μM. This compound is added at concentrations ranging from 0.125 to 128 μM and incubated with the pre-assembled WDR5/RbBP5/ASH2L complex at a final concentration of 0.5 μM for each protein for 2–5 min. Reactions are initiated by addition of the MLL1 protein at a final concentration of 0.5 μM and allowed to proceed for 30 min before preparing scintillation counting. To count samples, reactions are spotted on separate squares of P81 filter paper and precipitated by submerging in freshly prepared 50 mM sodium bicarbonate buffer with pH 9.0. After washing and drying, samples are vortexed in Ultima Gold scintillation fluid and counted. As a negative control, assays are performed using 0.5 μM MLL1/WDR5/RbBP5/ASH2L complex assembled with the non-interacting mutant, WDR5D107A.
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Growth inhibition assay | Cell viability |
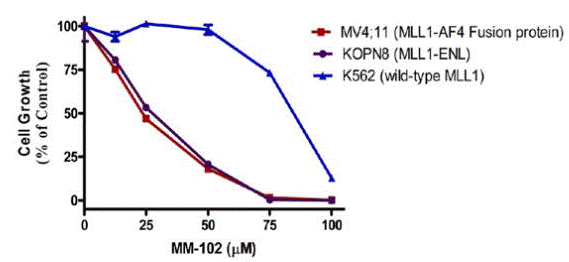
|
23210835 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































