research use only
GSK343 EZH2 Inhibitor
Cat.No.S7164
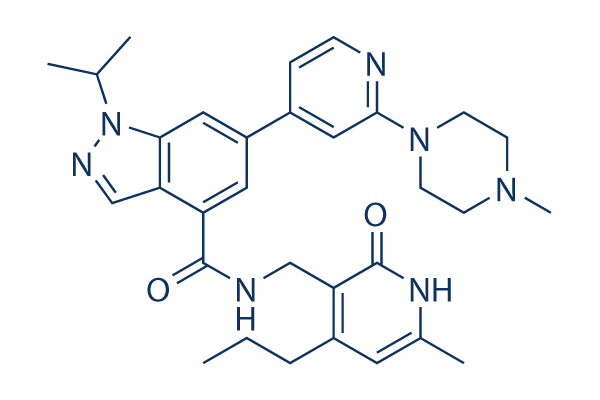
Chemical Structure
Molecular Weight: 541.69
Quality Control
| Related Targets | HDAC JAK BET Histone Methyltransferase PKC PARP HIF PRMT AMPK Histone Acetyltransferase |
|---|---|
| Other EZH2 Inhibitors | PF-06821497 GSK126 Tulmimetostat (CPI-0209) Valemetostat (DS-3201) Lirametostat (CPI-1205) CPI-169 SHR2554 EZH2/HSP90-IN-29 EBI-2511 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| 2D10 | Function Assay | 0.25-2.0 μM | 96 h | reduces total cellular trimethylated H3K27 in a time- and dose-dependent manner | 26041287 | |
| MDA-MB-231 | Growth Inhibition Assay | 0-20 μM | 72 h | shows cytotoxicity in a dose-dependent manner | 25203626 | |
| MDA-MB-231 | Function Assay | 10 μM | 72 h | reduces the level of H3K27-me3 | 25203626 | |
| MDA-MB-231 | Function Assay | 10 μM | 72 h | induces LC3-II accumulation | 25203626 | |
| MDA-MB-231 | Function Assay | 10 μM | 24 h | induces autophagy | 25203626 | |
| HepG2 | Growth Inhibition Assay | 0-20 μM | 72 h | shows cytotoxicity in a dose-dependent manner | 25203626 | |
| A549 | Growth Inhibition Assay | 0-20 μM | 72 h | shows cytotoxicity in a dose-dependent manner | 25203626 | |
| HepG2 | Function Assay | 10 μM | 24 h | induces autophagy | 25203626 | |
| A549 | Function Assay | 10 μM | 24 h | induces autophagy | 25203626 | |
| OVCAR10 | Growth Inhibition Assay | 1 μM | 0-12 d | DMSO | inhibits cell growth time dependently | 23759589 |
| UPN289 | Growth Inhibition Assay | 1 μM | 0-12 d | DMSO | inhibits cell growth time dependently | 23759589 |
| SKOV3 | Growth Inhibition Assay | 1 μM | 0-12 d | DMSO | inhibits cell growth time dependently | 23759589 |
| SKOV3 | Apoptosis Assay | 1 μM | 4 d | DMSO | induces apoptosis cultured in 3D conditions | 23759589 |
| HCC1806 | Function assay | 72 hrs | Inhibition of EZH2-mediated nuclear H3K27 methylation in human HCC1806 cells after 72 hrs by immunofluorescence analysis, IC50 = 0.174 μM. | 24900432 | ||
| LNCaP | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human LNCaP cells after 6 days by chemiluminescence analysis, IC50 = 2.9 μM. | 24900432 | ||
| DU145 | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human DU145 cells after 6 days by chemiluminescence analysis | 24900432 | ||
| SKBR3 | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human SKBR3 cells after 6 days by chemiluminescence analysis | 24900432 | ||
| PC3 | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human PC3 cells after 6 days by chemiluminescence analysis | 24900432 | ||
| ZR-75-1 | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human ZR-75-1 cells after 6 days by chemiluminescence analysis | 24900432 | ||
| HCC180 | Function assay | 6 days | Inhibition of EZH2-mediated proliferation of human HCC180 cells after 6 days by chemiluminescence analysis | 24900432 | ||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
Ethanol : 4 mg/mL
DMSO
: 1 mg/mL
(1.84 mM)
Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 541.69 | Formula | C31H39N7O2 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1346704-33-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CCCC1=C(C(=O)NC(=C1)C)CNC(=O)C2=C3C=NN(C3=CC(=C2)C4=CC(=NC=C4)N5CCN(CC5)C)C(C)C | ||
Mechanism of Action
| Features |
A chemical probe for the SGC epigenetics initiative. Potential use in a variety of solid tumors.
|
|---|---|
| Targets/IC50/Ki |
EZH2
(Cell-free assay) 4 nM
EZH1
(Cell-free assay) 240 nM
|
| In vitro |
GSK343 inhibits trimethylation of H3K27 (H3K27me3) with IC50 of 174 nM in HCC1806 breast cancer cells. This compound potently inhibits cell proliferation in breast cancer cells and prostate cancer cells, and the prostate cancer cell line LNCaP is the most sensitive to it, with IC50 of 2.9 μM. This chemical significantly suppresses the growth of EOC cells cultured in 3D matrigel extracellular matrix (ECM), which mimics the tumor microenvironment in vivo. In addition, it also induces apoptosis of EOC cells in 3D and significantly inhibits the invasion of EOC cells. |
| Kinase Assay |
In vitro biochemical assays against histone methyltransferases
|
|
Activity against EZH2 is assessed using 5 member PRC2 complex (Flag-EZH2, EED, SUZ12, AEBP2, RbAp48). The assay protocol may be summarized as follows: 10 mM stocks of compounds are prepared from solid in 100% DMSO. An 11 point serial dilution master plate is prepared in 384 well format (1:3 dilution, columns 6 and 18 were equal volume DMSO controls) and dispensed to assay ready plates using acoustic dispensing technology to create a 100 nL stamp of compound and DMSO controls. The assay additions consisted of equal volume additions of 10 nM EZH2 and the substrate solution (5 µg/mL HeLa nucleosomes and 0.25 µM [3H]-SAM) dispensed into assay plates using a multi-drop combi dispense. Reaction plates are incubated for 1 hr and quenched with an equal volume addition of 0.5 mg/mL PS-PEI Imaging Beads (RPNQ0098) containing 0.1 mM unlabeled SAM. The plates are sealed, dark adapted for 30 minutes, and a 5 minute endpoint luminescence image is acquired using a Viewlux imager. Plate statistics such as Z’ and signal to background as well as dose response curves are analyzed using Activity BaseXE. The in vitro biochemical activity of EZH1 is assessed as part of a 5 member PRC2 complex using a 384 well SPA assay identical to EZH2. Buffer components, reagent dispensing, compound plate preparation, quench conditions and data analysis are identical for EZH1 and EZH2 with final assay concentrations of 20 nM EZH1, 5 μM/mL HeLa nucleosomes and 0.25 μM [3H]-SAM. Further data analysis, pIC50 pivots and visualizations are enabled by TIBCO Spotfire. This compound is profiled at Reaction Biology Corp. (Malvern, PA) to assess inhibition in their panel of histone methyltransferase assays. Methyltransferase activity is assessed using HotSpot technology, a miniaturized radioisotope-based filter binding assay. Inhibitors are dissolved in dimethyl sulfoxide (DMSO) and tested at concentrations up to 100 uM with a final DMSO concentration of 2%. Buffer containing the methyltrasferase at the listed concentration and its preferred substrate as shown in the accompanying table is preincubated with this chemical for 10 min. Reactions are initiated by the addition of 1 uM S-adenosyl-L-[methyl-3H]methionine (SAM), allowed to incubate for 60 min at 30C followed by transfer to P81 filter-paper and PBS wash before detection.
|
|
| In vivo |
GSK343, a selective EZH2 inhibitor, inhibits phagocytosis. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
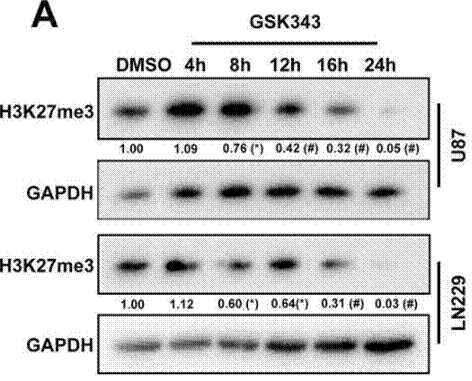
| Western blot | H3K27me3 EZH2 / EED / SUZ12 N-cadherin / Vimentin / Snail / Slug / MMP2 / MMP9 |
|
29228694 |
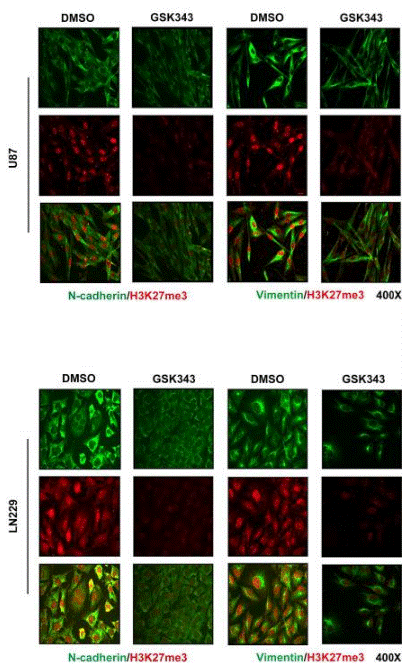
| Immunofluorescence | N-cadherin / H3K27me3 / Vimentin |
|
29228694 |
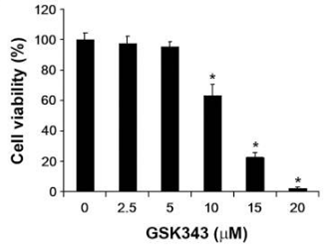
| Growth inhibition assay | Cell viability |
|
26973856 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































