research use only
GSK126 EZH2 inhibitor
Cat.No.S7061
Chemical Structure
Molecular Weight: 526.67
Quality Control
| Related Targets | HDAC JAK BET Histone Methyltransferase PKC PARP HIF PRMT AMPK Histone Acetyltransferase |
|---|---|
| Other EZH2 Inhibitors | PF-06821497 Tulmimetostat (CPI-0209) Valemetostat (DS-3201) GSK343 Lirametostat (CPI-1205) CPI-169 SHR2554 EZH2/HSP90-IN-29 EBI-2511 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human U87MG cells | Cytotoxic assay | 72 h | Cytotoxicity against human U87MG cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=28.5 μM. | 24767850 | ||
| human A549 cells | Cytotoxic assay | 72 h | Cytotoxicity against human A549 cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=18.7 μM. | 24767850 | ||
| human T98G cells | Cytotoxic assay | 72 h | Cytotoxicity against human T98G cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=12.6 μM. | 24767850 | ||
| human Daudi cells | Cytotoxic assay | 72 h | Cytotoxicity against human Daudi cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=11.2 μM. | 24767850 | ||
| human PC3 cells | Cytotoxic assay | 72 h | Cytotoxicity against human PC3 cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=9.4 μM. | 24767850 | ||
| human U2932 cells | Cytotoxic assay | 72 h | Cytotoxicity against human U2932 cells assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=6.7 μM. | 24767850 | ||
| human HeLa cells | Function assay | 72 h | Inhibition of EZH2 in human HeLa cells assessed as reduction in H3K27me3 levels incubated for 72 hrs by ELISA method, IC50=0.28 μM. | 26189078 | ||
| human Pfeiffer cells | Cytotoxic assay | 72 h | Cytotoxicity against human Pfeiffer cells expressing EZH2 A667G mutant assessed as growth inhibition after 72 hrs by WST-1 assay, GI50=0.18 μM. | 24767850 | ||
| infected SF9 cells | Binding affinity to EZH2 (unknown origin) expressed in baculovirus infected SF9 cells co-expressing SUZ12/EED/RbAp48 complex assessed as binding off-rate at 0.4 uM incubated for 20 mins by Q-TOF mass spectrometry | 27512831 | ||||
| A673 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | ||||
| DAOY cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for DAOY cells | 29435139 | ||||
| Saos-2 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Saos-2 cells | 29435139 | ||||
| BT-37 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | ||||
| RD cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | ||||
| SK-N-SH cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | ||||
| BT-12 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-12 cells | 29435139 | ||||
| MG 63 (6-TG R) cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | ||||
| NB1643 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | ||||
| OHS-50 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | ||||
| Rh41 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh41 cells | 29435139 | ||||
| SK-N-MC cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | ||||
| LAN-5 cells | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 5 mg/mL
(9.49 mM)
Ethanol : 4 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 526.67 | Formula | C31H38N6O2 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1346574-57-9 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | GSK2816126A, GSK2816126 | Smiles | CCC(C)N1C=C(C2=C(C=C(C=C21)C3=CN=C(C=C3)N4CCNCC4)C(=O)NCC5=C(C=C(NC5=O)C)C)C | ||
Mechanism of Action
| Targets/IC50/Ki |
EZH2
(Cell-free assay) 9.9 nM
|
|---|---|
| In vitro |
In vitro, GSK126 most potently inhibits H3K27me3, followed by H3K27me2 in both EZH2 wild-type and mutant DLBCL cell lines. This compound also effectively inhibits the proliferation of EZH2 mutant DLBCL cell lines, and induces transcriptional activation of EZH2 target genes in sensitive cell lines. In A687V EZH2-mutant cells, this treatment results in a global decrease in H3K27me3, robust gene activation, caspase activation, and decreased proliferation. In parental H2087 cells, it inhibits the expression of VEGF-A and phosphorylated Ser(473)-AKT, and thus causes the inhibition of cell proliferation, migration and metastasis.
|
| Kinase Assay |
EZH2 assay
|
|
The five-member PRC2 complex (Flag–EZH2, EED, SUZ12, AEBP2, RbAp48) containing either wild-type or mutant EZH2 is prepared. GSK126 is dissolved in DMSO and tested at concentrations of 0.6 nM to 300 nM with a final DMSO concentration of 2.5%. In contrast to wild-type EZH2 which prefers H3K27me0 as a substrate in vitro, EZH2 Y641 mutants prefer H3K27me2 and have little activity with H3K27me0 or H3K27me1. The A677G mutant is distinct from both the wild-type and Y641 mutant forms of EZH2 in that it efficiently methylates H3K27me0, H3K27me1, and H3K27me2; therefore, histone H3 peptides (residues 21–44; 10 μM final) with either K27me0 (wild type, A677G EZH2), K27me1 (A677G EZH2), or K27me2 (A677G, Y641N, Y641C, Y641H, Y641S and Y641F EZH2) are used as methyltransferase substrates. This compound is added to plates followed by addition of 6 nM EZH2 complex and peptide. As the potency of this chemical is at or near the tight binding limit of an assay run at [SAM] = Km, IC50 values are measured at a high concentration of the competitive substrate SAM relative to its Km (7.5 μM SAM where the SAM Km is 0.3 μM). Under these conditions, the contribution from the enzyme concentration becomes relatively small and accurate estimates of Ki can be calculated. Reactions are initiated with [3H]-SAM, incubated for 30 min, quenched with the addition of 500-fold excess unlabelled SAM, and the methylated product peptide is captured on phosphocellulose filters according to the vendor supplied protocol for MSPH Multiscreen plates. Plates are read on a TopCount after adding 20 μL of Microscint-20 cocktail. Apparent Ki values are calculated using the Cheng–Prusoff relationship for a competitive inhibitor. IC50=Ki (1+[S]/Km)+[E]/2, where E is the enzyme and S is the substrate.
|
|
| In vivo |
In mice bearing KARPAS-422 and Pfeiffer xenografts, GSK126 (150 mg/kg/d, i.p.) decreases global H3K27me3, increases gene expression, and thus causes marked tumour regression.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
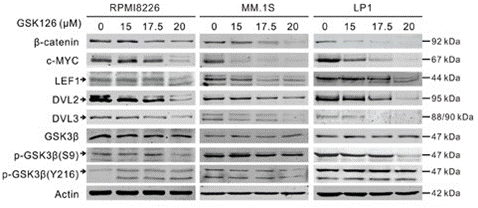
| Western blot | β-catenin / c-Myc / LEF1 / DVL2 / DVL3 / p-GSK3β XIAP / Survivin / MCL-1 / BID / BIM / BAX / BCL-xl/ Bcl-2 H3K27Me3 / EZH2 |
|
27926488 |
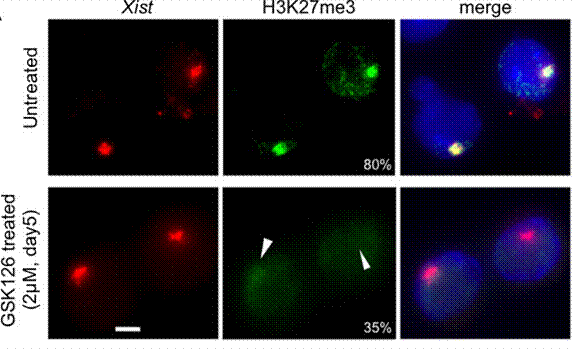
| Immunofluorescence | H3K27me3 |
|
25053977 |
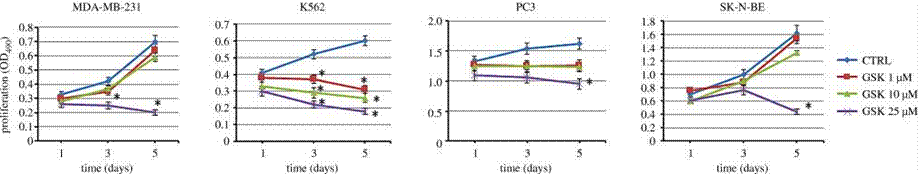
| Growth inhibition assay | Cell proliferation Cell viability |
|
29685965 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT02082977 | Terminated | Cancer|Neoplasms |
GlaxoSmithKline |
April 24 2014 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
Could you please suggest a vehicle for in vivo uses of this compound without oil?
Answer:
It could be dissolved in 4% DMSO+30% PEG 300+ddH2O (0.5mg/ml).
Question 2:
Does this compound require an activation step to be functional? For example, an acidic or basic environment.
Answer:
It does not require an activation step to be functional.






































