research use only
CHIR-99021 (Laduviglusib) GSK-3 inhibitor
Cat.No.S1263
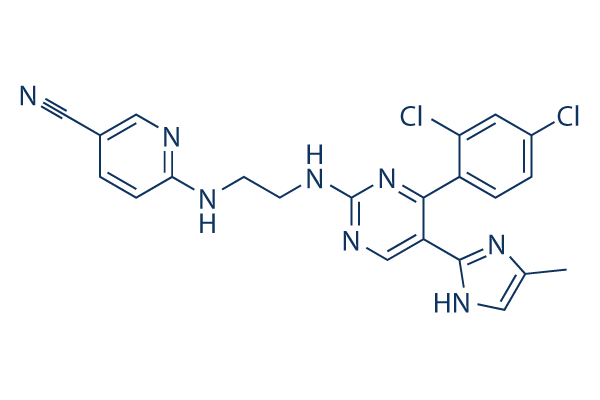
Chemical Structure
Molecular Weight: 465.34
Quality Control
Products Often Used Together with CHIR-99021 (Laduviglusib)
| Related Targets | PI3K Akt mTOR ATM/ATR DNA-PK AMPK PDPK1 PTEN PP2A PDK |
|---|---|
| Other GSK-3 Inhibitors | Laduviglusib (CHIR-99021) Hydrochloride SB216763 CHIR-98014 TWS119 GSK-3 Inhibitor IX (BIO) LY2090314 Tideglusib SB415286 AR-A014418 IM-12 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| LOUCY | Growth Inhibition Assay | IC50=182.688 μM | SANGER | |||
| SN12C | Growth Inhibition Assay | IC50=182.829 μM | SANGER | |||
| A427 | Growth Inhibition Assay | IC50=184.631 μM | SANGER | |||
| DOK | Growth Inhibition Assay | IC50=184.643 μM | SANGER | |||
| MDA-MB-415 | Growth Inhibition Assay | IC50=184.651 μM | SANGER | |||
| MRK-nu-1 | Growth Inhibition Assay | IC50=184.972 μM | SANGER | |||
| LB2241-RCC | Growth Inhibition Assay | IC50=185.145 μM | SANGER | |||
| NCI-H1563 | Growth Inhibition Assay | IC50=185.209 μM | SANGER | |||
| MDA-MB-453 | Growth Inhibition Assay | IC50=186.941 μM | SANGER | |||
| MONO-MAC-6 | Growth Inhibition Assay | IC50=187.392 μM | SANGER | |||
| CAPAN-1 | Growth Inhibition Assay | IC50=187.453 μM | SANGER | |||
| SK-LU-1 | Growth Inhibition Assay | IC50=188.428 μM | SANGER | |||
| NCI-H1299 | Growth Inhibition Assay | IC50=188.719 μM | SANGER | |||
| KG-1 | Growth Inhibition Assay | IC50=189.798 μM | SANGER | |||
| ETK-1 | Growth Inhibition Assay | IC50=190.181 μM | SANGER | |||
| NCI-H838 | Growth Inhibition Assay | IC50=190.34 μM | SANGER | |||
| HCE-4 | Growth Inhibition Assay | IC50=190.992 μM | SANGER | |||
| A2780 | Growth Inhibition Assay | IC50=192.891 μM | SANGER | |||
| COLO-668 | Growth Inhibition Assay | IC50=193.157 μM | SANGER | |||
| HGC-27 | Growth Inhibition Assay | IC50=195.538 μM | SANGER | |||
| SW1088 | Growth Inhibition Assay | IC50=195.677 μM | SANGER | |||
| KOSC-2 | Growth Inhibition Assay | IC50=195.818 μM | SANGER | |||
| SW780 | Growth Inhibition Assay | IC50=196.734 μM | SANGER | |||
| TE-1 | Growth Inhibition Assay | IC50=196.26 μM | SANGER | |||
| 22RV1 | Growth Inhibition Assay | IC50=197.424 μM | SANGER | |||
| IA-LM | Growth Inhibition Assay | IC50=197.822 μM | SANGER | |||
| TE-5 | Growth Inhibition Assay | IC50=198.033 μM | SANGER | |||
| JVM-3 | Growth Inhibition Assay | IC50=198.419 μM | SANGER | |||
| HT55 | Growth Inhibition Assay | IC50=199.192 μM | SANGER | |||
| QIMR-WIL | Growth Inhibition Assay | IC50=200.116 μM | SANGER | |||
| SJRH30 | Growth Inhibition Assay | IC50=200.515 μM | SANGER | |||
| COLO-679 | Growth Inhibition Assay | IC50=200.859 μM | SANGER | |||
| MFM-223 | Growth Inhibition Assay | IC50=201.587 μM | SANGER | |||
| MDA-MB-231 | Growth Inhibition Assay | IC50=200.902 μM | SANGER | |||
| MFE-280 | Growth Inhibition Assay | IC50=201.961 μM | SANGER | |||
| LAMA-84 | Growth Inhibition Assay | IC50=202.073 μM | SANGER | |||
| LS-1034 | Growth Inhibition Assay | IC50=202.553 μM | SANGER | |||
| NCI-H345 | Growth Inhibition Assay | IC50=203.027 μM | SANGER | |||
| KYSE-70 | Growth Inhibition Assay | IC50=203.879 μM | SANGER | |||
| EGI-1 | Growth Inhibition Assay | IC50=204.833 μM | SANGER | |||
| RPMI-2650 | Growth Inhibition Assay | IC50=205.467 μM | SANGER | |||
| NCI-H1651 | Growth Inhibition Assay | IC50=206.73 μM | SANGER | |||
| HPAF-II | Growth Inhibition Assay | IC50=209.671 μM | SANGER | |||
| NUGC-3 | Growth Inhibition Assay | IC50=209.678 μM | SANGER | |||
| DJM-1 | Growth Inhibition Assay | IC50=209.897 μM | SANGER | |||
| MHH-PREB-1 | Growth Inhibition Assay | IC50=210.381 μM | SANGER | |||
| EW-18 | Growth Inhibition Assay | IC50=210.763 μM | SANGER | |||
| BT-20 | Growth Inhibition Assay | IC50=210.986 μM | SANGER | |||
| A431 | Growth Inhibition Assay | IC50=211.299 μM | SANGER | |||
| TCCSUP | Growth Inhibition Assay | IC50=211.533 μM | SANGER | |||
| HO-1-N-1 | Growth Inhibition Assay | IC50=212.894 μM | SANGER | |||
| NCI-H69 | Growth Inhibition Assay | IC50=214.185 μM | SANGER | |||
| ES7 | Growth Inhibition Assay | IC50=214.878 μM | SANGER | |||
| C3A | Growth Inhibition Assay | IC50=215.575 μM | SANGER | |||
| UACC-257 | Growth Inhibition Assay | IC50=217.43 μM | SANGER | |||
| DMS-153 | Growth Inhibition Assay | IC50=217.836 μM | SANGER | |||
| LB771-HNC | Growth Inhibition Assay | IC50=217.893 μM | SANGER | |||
| HOS | Growth Inhibition Assay | IC50=218.9 μM | SANGER | |||
| THP-1 | Growth Inhibition Assay | IC50=219.606 μM | SANGER | |||
| SCC-4 | Growth Inhibition Assay | IC50=220.353 μM | SANGER | |||
| SCC-3 | Growth Inhibition Assay | IC50=220.577 μM | SANGER | |||
| BC-1 | Growth Inhibition Assay | IC50=220.799 μM | SANGER | |||
| NCI-H1618 | Growth Inhibition Assay | IC50=221.018 μM | SANGER | |||
| CaR-1 | Growth Inhibition Assay | IC50=221.251 μM | SANGER | |||
| NCI-H1355 | Growth Inhibition Assay | IC50=221.377 μM | SANGER | |||
| NCI-H1993 | Growth Inhibition Assay | IC50=222.239 μM | SANGER | |||
| NCI-H1623 | Growth Inhibition Assay | IC50=222.389 μM | SANGER | |||
| AGS | Growth Inhibition Assay | IC50=222.523 μM | SANGER | |||
| SAS | Growth Inhibition Assay | IC50=223.796 μM | SANGER | |||
| HCC1937 | Growth Inhibition Assay | IC50=224.389 μM | SANGER | |||
| NCI-H209 | Growth Inhibition Assay | IC50=226.53 μM | SANGER | |||
| NCI-H322M | Growth Inhibition Assay | IC50=228.019 μM | SANGER | |||
| CHL-1 | Growth Inhibition Assay | IC50=228.13 μM | SANGER | |||
| NCI-H630 | Growth Inhibition Assay | IC50=232.897 μM | SANGER | |||
| 5637 | Growth Inhibition Assay | IC50=233.003 μM | SANGER | |||
| MSTO-211H | Growth Inhibition Assay | IC50=233.276 μM | SANGER | |||
| YAPC | Growth Inhibition Assay | IC50=234.253 μM | SANGER | |||
| SW1783 | Growth Inhibition Assay | IC50=234.41 μM | SANGER | |||
| GB-1 | Growth Inhibition Assay | IC50=235.177 μM | SANGER | |||
| NCI-H520 | Growth Inhibition Assay | IC50=235.741 μM | SANGER | |||
| NOMO-1 | Growth Inhibition Assay | IC50=236.02 μM | SANGER | |||
| NCCIT | Growth Inhibition Assay | IC50=236.873 μM | SANGER | |||
| SW954 | Growth Inhibition Assay | IC50=237.868 μM | SANGER | |||
| MOLT-4 | Growth Inhibition Assay | IC50=241.209 μM | SANGER | |||
| EB2 | Growth Inhibition Assay | IC50=241.907 μM | SANGER | |||
| IPC-298 | Growth Inhibition Assay | IC50=242.072 μM | SANGER | |||
| GP5d | Growth Inhibition Assay | IC50=242.616 μM | SANGER | |||
| HN | Growth Inhibition Assay | IC50=242.991 μM | SANGER | |||
| BFTC-909 | Growth Inhibition Assay | IC50=243.118 μM | SANGER | |||
| ES5 | Growth Inhibition Assay | IC50=245.038 μM | SANGER | |||
| SK-HEP-1 | Growth Inhibition Assay | IC50=246.043 μM | SANGER | |||
| T47D | Growth Inhibition Assay | IC50=247.013 μM | SANGER | |||
| KYSE-510 | Growth Inhibition Assay | IC50=247.517 μM | SANGER | |||
| LC-1F | Growth Inhibition Assay | IC50=248.156 μM | SANGER | |||
| NCI-H1693 | Growth Inhibition Assay | IC50=249.495 μM | SANGER | |||
| COR-L23 | Growth Inhibition Assay | IC50=250.412 μM | SANGER | |||
| NCI-H2228 | Growth Inhibition Assay | IC50=250.864 μM | SANGER | |||
| NCI-H460 | Growth Inhibition Assay | IC50=251.417 μM | SANGER | |||
| TGBC1TKB | Growth Inhibition Assay | IC50=252.875 μM | SANGER | |||
| NCI-H1304 | Growth Inhibition Assay | IC50=253.156 μM | SANGER | |||
| CAL-27 | Growth Inhibition Assay | IC50=253.485 μM | SANGER | |||
| NCI-H187 | Growth Inhibition Assay | IC50=256.76 μM | SANGER | |||
| HSC-2 | Growth Inhibition Assay | IC50=258.584 μM | SANGER | |||
| CAL-39 | Growth Inhibition Assay | IC50=260.272 μM | SANGER | |||
| HuH-7 | Growth Inhibition Assay | IC50=260.996 μM | SANGER | |||
| MMAC-SF | Growth Inhibition Assay | IC50=261.192 μM | SANGER | |||
| VA-ES-BJ | Growth Inhibition Assay | IC50=261.918 μM | SANGER | |||
| A673 | Growth Inhibition Assay | IC50=267.025 μM | SANGER | |||
| LU-165 | Growth Inhibition Assay | IC50=267.941 μM | SANGER | |||
| CAL-12T | Growth Inhibition Assay | IC50=268.366 μM | SANGER | |||
| RD | Growth Inhibition Assay | IC50=269.119 μM | SANGER | |||
| MDA-MB-468 | Growth Inhibition Assay | IC50=269.333 μM | SANGER | |||
| LN-405 | Growth Inhibition Assay | IC50=269.708 μM | SANGER | |||
| MHH-CALL-2 | Growth Inhibition Assay | IC50=269.92 μM | SANGER | |||
| ES3 | Growth Inhibition Assay | IC50=270.358 μM | SANGER | |||
| SK-MEL-3 | Growth Inhibition Assay | IC50=275.457 μM | SANGER | |||
| TE-10 | Growth Inhibition Assay | IC50=276.439 μM | SANGER | |||
| BEN | Growth Inhibition Assay | IC50=276.461 μM | SANGER | |||
| NB69 | Growth Inhibition Assay | IC50=277.405 μM | SANGER | |||
| CAMA-1 | Growth Inhibition Assay | IC50=278.279 μM | SANGER | |||
| NCI-H1792 | Growth Inhibition Assay | IC50=279.187 μM | SANGER | |||
| UACC-893 | Growth Inhibition Assay | IC50=280.54 μM | SANGER | |||
| TGW | Growth Inhibition Assay | IC50=283.058 μM | SANGER | |||
| SH-4 | Growth Inhibition Assay | IC50=283.123 μM | SANGER | |||
| NCI-H2009 | Growth Inhibition Assay | IC50=283.953 μM | SANGER | |||
| T84 | Growth Inhibition Assay | IC50=284.665 μM | SANGER | |||
| EW-7 | Growth Inhibition Assay | IC50=285.232 μM | SANGER | |||
| NCI-H1882 | Growth Inhibition Assay | IC50=286.035 μM | SANGER | |||
| GMS-10 | Growth Inhibition Assay | IC50=287.183 μM | SANGER | |||
| HC-1 | Growth Inhibition Assay | IC50=290.602 μM | SANGER | |||
| ABC-1 | Growth Inhibition Assay | IC50=291.069 μM | SANGER | |||
| HuP-T3 | Growth Inhibition Assay | IC50=292.146 μM | SANGER | |||
| HT-1197 | Growth Inhibition Assay | IC50=292.879 μM | SANGER | |||
| SHP-77 | Growth Inhibition Assay | IC50=295.597 μM | SANGER | |||
| NCI-H524 | Growth Inhibition Assay | IC50=295.769 μM | SANGER | |||
| SW626 | Growth Inhibition Assay | IC50=296.115 μM | SANGER | |||
| NCI-H2347 | Growth Inhibition Assay | IC50=296.425 μM | SANGER | |||
| U251 | Growth Inhibition Assay | IC50=298.438 μM | SANGER | |||
| RL95-2 | Growth Inhibition Assay | IC50=300.247 μM | SANGER | |||
| C2BBe1 | Growth Inhibition Assay | IC50=300.691 μM | SANGER | |||
| HLE | Growth Inhibition Assay | IC50=301.818 μM | SANGER | |||
| COLO-680N | Growth Inhibition Assay | IC50=302.893 μM | SANGER | |||
| PLC-PRF-5 | Growth Inhibition Assay | IC50=304.907 μM | SANGER | |||
| SCC-15 | Growth Inhibition Assay | IC50=306.017 μM | SANGER | |||
| ECC12 | Growth Inhibition Assay | IC50=310.254 μM | SANGER | |||
| A388 | Growth Inhibition Assay | IC50=312.022 μM | SANGER | |||
| ES1 | Growth Inhibition Assay | IC50=312.189 μM | SANGER | |||
| J-RT3-T3-5 | Growth Inhibition Assay | IC50=313.521 μM | SANGER | |||
| SNU-475 | Growth Inhibition Assay | IC50=314.091 μM | SANGER | |||
| 647-V | Growth Inhibition Assay | IC50=314.673 μM | SANGER | |||
| IMR-5 | Growth Inhibition Assay | IC50=315.38 μM | SANGER | |||
| EW-24 | Growth Inhibition Assay | IC50=320.07 μM | SANGER | |||
| CAL-51 | Growth Inhibition Assay | IC50=321.403 μM | SANGER | |||
| HD-MY-Z | Growth Inhibition Assay | IC50=321.409 μM | SANGER | |||
| EW-13 | Growth Inhibition Assay | IC50=322.424 μM | SANGER | |||
| SK-N-FI | Growth Inhibition Assay | IC50=322.452 μM | SANGER | |||
| BHY | Growth Inhibition Assay | IC50=323.175 μM | SANGER | |||
| OCUB-M | Growth Inhibition Assay | IC50=323.434 μM | SANGER | |||
| HCC1954 | Growth Inhibition Assay | IC50=323.582 μM | SANGER | |||
| SK-MEL-24 | Growth Inhibition Assay | IC50=324.111 μM | SANGER | |||
| NTERA-S-cl-D1 | Growth Inhibition Assay | IC50=326.489 μM | SANGER | |||
| SK-MEL-30 | Growth Inhibition Assay | IC50=326.738 μM | SANGER | |||
| LS-123 | Growth Inhibition Assay | IC50=328.164 μM | SANGER | |||
| SF268 | Growth Inhibition Assay | IC50=330.265 μM | SANGER | |||
| BT-474 | Growth Inhibition Assay | IC50=332.418 μM | SANGER | |||
| NCI-H1522 | Growth Inhibition Assay | IC50=334.339 μM | SANGER | |||
| LC-2-ad | Growth Inhibition Assay | IC50=336.591 μM | SANGER | |||
| LAN-6 | Growth Inhibition Assay | IC50=336.977 μM | SANGER | |||
| ARH-77 | Growth Inhibition Assay | IC50=337.32 μM | SANGER | |||
| HuO9 | Growth Inhibition Assay | IC50=337.47 μM | SANGER | |||
| MLMA | Growth Inhibition Assay | IC50=338.155 μM | SANGER | |||
| Raji | Growth Inhibition Assay | IC50=338.38 μM | SANGER | |||
| GT3TKB | Growth Inhibition Assay | IC50=338.412 μM | SANGER | |||
| NCI-H441 | Growth Inhibition Assay | IC50=341.418 μM | SANGER | |||
| NCI-H2030 | Growth Inhibition Assay | IC50=341.609 μM | SANGER | |||
| VM-CUB-1 | Growth Inhibition Assay | IC50=342.636 μM | SANGER | |||
| KP-4 | Growth Inhibition Assay | IC50=343.733 μM | SANGER | |||
| PF-382 | Growth Inhibition Assay | IC50=344.422 μM | SANGER | |||
| NB13 | Growth Inhibition Assay | IC50=345.58 μM | SANGER | |||
| D-502MG | Growth Inhibition Assay | IC50=345.812 μM | SANGER | |||
| CAL-54 | Growth Inhibition Assay | IC50=345.879 μM | SANGER | |||
| DOHH-2 | Growth Inhibition Assay | IC50=346.65 μM | SANGER | |||
| SK-PN-DW | Growth Inhibition Assay | IC50=347.426 μM | SANGER | |||
| NCI-H720 | Growth Inhibition Assay | IC50=349.846 μM | SANGER | |||
| SW1990 | Growth Inhibition Assay | IC50=351.28 μM | SANGER | |||
| NCI-H2196 | Growth Inhibition Assay | IC50=352.624 μM | SANGER | |||
| P30-OHK | Growth Inhibition Assay | IC50=353.063 μM | SANGER | |||
| NCI-H358 | Growth Inhibition Assay | IC50=355.356 μM | SANGER | |||
| CW-2 | Growth Inhibition Assay | IC50=356.529 μM | SANGER | |||
| COLO-678 | Growth Inhibition Assay | IC50=357.863 μM | SANGER | |||
| NCI-H2126 | Growth Inhibition Assay | IC50=358.521 μM | SANGER | |||
| DMS-53 | Growth Inhibition Assay | IC50=358.711 μM | SANGER | |||
| NCI-H1838 | Growth Inhibition Assay | IC50=359.987 μM | SANGER | |||
| SJSA-1 | Growth Inhibition Assay | IC50=360.939 μM | SANGER | |||
| ChaGo-K-1 | Growth Inhibition Assay | IC50=363.924 μM | SANGER | |||
| IST-SL2 | Growth Inhibition Assay | IC50=372.926 μM | SANGER | |||
| U-266 | Growth Inhibition Assay | IC50=373.132 μM | SANGER | |||
| P31-FUJ | Growth Inhibition Assay | IC50=376.964 μM | SANGER | |||
| HCC2157 | Growth Inhibition Assay | IC50=378.161 μM | SANGER | |||
| COR-L88 | Growth Inhibition Assay | IC50=383.677 μM | SANGER | |||
| NCI-H596 | Growth Inhibition Assay | IC50=385.34 μM | SANGER | |||
| GOTO | Growth Inhibition Assay | IC50=390.641 μM | SANGER | |||
| NCI-H719 | Growth Inhibition Assay | IC50=391.893 μM | SANGER | |||
| MN-60 | Growth Inhibition Assay | IC50=393.948 μM | SANGER | |||
| IST-SL1 | Growth Inhibition Assay | IC50=394.406 μM | SANGER | |||
| TE-6 | Growth Inhibition Assay | IC50=397.254 μM | SANGER | |||
| SW962 | Growth Inhibition Assay | IC50=398.69 μM | SANGER | |||
| FADU | Growth Inhibition Assay | IC50=403.083 μM | SANGER | |||
| ECC4 | Growth Inhibition Assay | IC50=408.458 μM | SANGER | |||
| MES-SA | Growth Inhibition Assay | IC50=409.98 μM | SANGER | |||
| NCI-H716 | Growth Inhibition Assay | IC50=410.075 μM | SANGER | |||
| GR-ST | Growth Inhibition Assay | IC50=410.497 μM | SANGER | |||
| KY821 | Growth Inhibition Assay | IC50=414.086 μM | SANGER | |||
| NCI-H2141 | Growth Inhibition Assay | IC50=424.388 μM | SANGER | |||
| MDA-MB-134-VI | Growth Inhibition Assay | IC50=431.033 μM | SANGER | |||
| RH-1 | Growth Inhibition Assay | IC50=435.09 μM | SANGER | |||
| AM-38 | Growth Inhibition Assay | IC50=438.54 μM | SANGER | |||
| BL-70 | Growth Inhibition Assay | IC50=439.429 μM | SANGER | |||
| COLO-684 | Growth Inhibition Assay | IC50=448.888 μM | SANGER | |||
| DMS-79 | Growth Inhibition Assay | IC50=449.68 μM | SANGER | |||
| EW-1 | Growth Inhibition Assay | IC50=452.888 μM | SANGER | |||
| MS-1 | Growth Inhibition Assay | IC50=454.385 μM | SANGER | |||
| A4-Fuk | Growth Inhibition Assay | IC50=454.507 μM | SANGER | |||
| TALL-1 | Growth Inhibition Assay | IC50=455.297 μM | SANGER | |||
| SCLC-21H | Growth Inhibition Assay | IC50=455.699 μM | SANGER | |||
| KMS-12-PE | Growth Inhibition Assay | IC50=456.822 μM | SANGER | |||
| TE-441-T | Growth Inhibition Assay | IC50=457.186 μM | SANGER | |||
| A3-KAW | Growth Inhibition Assay | IC50=460.841 μM | SANGER | |||
| NCI-H1092 | Growth Inhibition Assay | IC50=463.943 μM | SANGER | |||
| COR-L279 | Growth Inhibition Assay | IC50=464.847 μM | SANGER | |||
| NCI-SNU-5 | Growth Inhibition Assay | IC50=472.28 μM | SANGER | |||
| JAR | Growth Inhibition Assay | IC50=474.788 μM | SANGER | |||
| NCI-H446 | Growth Inhibition Assay | IC50=477.08 μM | SANGER | |||
| HDLM-2 | Growth Inhibition Assay | IC50=480.869 μM | SANGER | |||
| D-283MED | Growth Inhibition Assay | IC50=488.924 μM | SANGER | |||
| NCI-H1395 | Growth Inhibition Assay | IC50=489.004 μM | SANGER | |||
| RPMI-6666 | Growth Inhibition Assay | IC50=491.398 μM | SANGER | |||
| CPC-N | Growth Inhibition Assay | IC50=502.063 μM | SANGER | |||
| EC-GI-10 | Growth Inhibition Assay | IC50=502.169 μM | SANGER | |||
| CA46 | Growth Inhibition Assay | IC50=504.407 μM | SANGER | |||
| EHEB | Growth Inhibition Assay | IC50=507.208 μM | SANGER | |||
| RPMI-8402 | Growth Inhibition Assay | IC50=510.021 μM | SANGER | |||
| DU-4475 | Growth Inhibition Assay | IC50=511.101 μM | SANGER | |||
| P12-ICHIKAWA | Growth Inhibition Assay | IC50=511.416 μM | SANGER | |||
| NCI-H1694 | Growth Inhibition Assay | IC50=512.439 μM | SANGER | |||
| NCI-H2171 | Growth Inhibition Assay | IC50=512.705 μM | SANGER | |||
| Mo-T | Growth Inhibition Assay | IC50=515.116 μM | SANGER | |||
| HL-60 | Growth Inhibition Assay | IC50=522.924 μM | SANGER | |||
| NCI-H1436 | Growth Inhibition Assay | IC50=523.064 μM | SANGER | |||
| NCI-H510A | Growth Inhibition Assay | IC50=524.696 μM | SANGER | |||
| NCI-H2081 | Growth Inhibition Assay | IC50=525.513 μM | SANGER | |||
| NB5 | Growth Inhibition Assay | IC50=526.43 μM | SANGER | |||
| NCI-H2227 | Growth Inhibition Assay | IC50=530.254 μM | SANGER | |||
| HSC-4 | Growth Inhibition Assay | IC50=539.293 μM | SANGER | |||
| RL | Growth Inhibition Assay | IC50=542.442 μM | SANGER | |||
| EVSA-T | Growth Inhibition Assay | IC50=543.684 μM | SANGER | |||
| NCI-H889 | Growth Inhibition Assay | IC50=547.23 μM | SANGER | |||
| COLO-320-HSR | Growth Inhibition Assay | IC50=551.35 μM | SANGER | |||
| EW-11 | Growth Inhibition Assay | IC50=552.399 μM | SANGER | |||
| UACC-812 | Growth Inhibition Assay | IC50=559.391 μM | SANGER | |||
| CAL-148 | Growth Inhibition Assay | IC50=565.089 μM | SANGER | |||
| K052 | Growth Inhibition Assay | IC50=579.213 μM | SANGER | |||
| L-428 | Growth Inhibition Assay | IC50=600.671 μM | SANGER | |||
| LP-1 | Growth Inhibition Assay | IC50=603.587 μM | SANGER | |||
| ML-2 | Growth Inhibition Assay | IC50=606.579 μM | SANGER | |||
| KARPAS-45 | Growth Inhibition Assay | IC50=618.221 μM | SANGER | |||
| HT | Growth Inhibition Assay | IC50=622.785 μM | SANGER | |||
| NCI-H226 | Growth Inhibition Assay | IC50=628.353 μM | SANGER | |||
| L-363 | Growth Inhibition Assay | IC50=648.064 μM | SANGER | |||
| IM-9 | Growth Inhibition Assay | IC50=651.541 μM | SANGER | |||
| TUR | Growth Inhibition Assay | IC50=671.322 μM | SANGER | |||
| NCI-SNU-16 | Growth Inhibition Assay | IC50=680.462 μM | SANGER | |||
| NCI-SNU-1 | Growth Inhibition Assay | IC50=690.172 μM | SANGER | |||
| U-87-MG | Growth Inhibition Assay | IC50=694.585 μM | SANGER | |||
| KM-H2 | Growth Inhibition Assay | IC50=699.059 μM | SANGER | |||
| DEL | Growth Inhibition Assay | IC50=700.838 μM | SANGER | |||
| KARPAS-422 | Growth Inhibition Assay | IC50=789.315 μM | SANGER | |||
| BC-3 | Growth Inhibition Assay | IC50=805.203 μM | SANGER | |||
| NCI-H1155 | Growth Inhibition Assay | IC50=818.059 μM | SANGER | |||
| NCI-H1581 | Growth Inhibition Assay | IC50=900.231 μM | SANGER | |||
| ONS-76 | Growth Inhibition Assay | IC50=182.387 μM | SANGER | |||
| SBC-5 | Growth Inhibition Assay | IC50=181.934 μM | SANGER | |||
| GI-ME-N | Growth Inhibition Assay | IC50=181.672 μM | SANGER | |||
| PSN1 | Growth Inhibition Assay | IC50=181.66 μM | SANGER | |||
| NCI-H522 | Growth Inhibition Assay | IC50=181.059 μM | SANGER | |||
| SF126 | Growth Inhibition Assay | IC50=179.432 μM | SANGER | |||
| NB10 | Growth Inhibition Assay | IC50=177.846 μM | SANGER | |||
| SK-NEP-1 | Growth Inhibition Assay | IC50=177.806 μM | SANGER | |||
| COLO-741 | Growth Inhibition Assay | IC50=177.561 μM | SANGER | |||
| EW-3 | Growth Inhibition Assay | IC50=176.555 μM | SANGER | |||
| YT | Growth Inhibition Assay | IC50=176.399 μM | SANGER | |||
| LNCaP-Clone-FGC | Growth Inhibition Assay | IC50=175.861 μM | SANGER | |||
| SW756 | Growth Inhibition Assay | IC50=174.863 μM | SANGER | |||
| ME-180 | Growth Inhibition Assay | IC50=174.016 μM | SANGER | |||
| HH | Growth Inhibition Assay | IC50=173.848 μM | SANGER | |||
| HuO-3N1 | Growth Inhibition Assay | IC50=172.273 μM | SANGER | |||
| SW620 | Growth Inhibition Assay | IC50=171.535 μM | SANGER | |||
| HUTU-80 | Growth Inhibition Assay | IC50=171.032 μM | SANGER | |||
| Daoy | Growth Inhibition Assay | IC50=169.891 μM | SANGER | |||
| KYSE-150 | Growth Inhibition Assay | IC50=169.636 μM | SANGER | |||
| CP66-MEL | Growth Inhibition Assay | IC50=168.415 μM | SANGER | |||
| Calu-3 | Growth Inhibition Assay | IC50=168.311 μM | SANGER | |||
| JVM-2 | Growth Inhibition Assay | IC50=167.451 μM | SANGER | |||
| OVCAR-5 | Growth Inhibition Assay | IC50=168.309 μM | SANGER | |||
| KINGS-1 | Growth Inhibition Assay | IC50=167.354 μM | SANGER | |||
| EFO-21 | Growth Inhibition Assay | IC50=165.45 μM | SANGER | |||
| Ca9-22 | Growth Inhibition Assay | IC50=164.321 μM | SANGER | |||
| AU565 | Growth Inhibition Assay | IC50=163.64 μM | SANGER | |||
| CHP-212 | Growth Inhibition Assay | IC50=163.626 μM | SANGER | |||
| CAL-62 | Growth Inhibition Assay | IC50=162.613 μM | SANGER | |||
| EFO-27 | Growth Inhibition Assay | IC50=160.584 μM | SANGER | |||
| NCI-H2052 | Growth Inhibition Assay | IC50=159.583 μM | SANGER | |||
| ES4 | Growth Inhibition Assay | IC50=159.396 μM | SANGER | |||
| HCC70 | Growth Inhibition Assay | IC50=159.242 μM | SANGER | |||
| HCC38 | Growth Inhibition Assay | IC50=157.369 μM | SANGER | |||
| DG-75 | Growth Inhibition Assay | IC50=156.694 μM | SANGER | |||
| HCC1419 | Growth Inhibition Assay | IC50=156.077 μM | SANGER | |||
| CAL-120 | Growth Inhibition Assay | IC50=155.821 μM | SANGER | |||
| DK-MG | Growth Inhibition Assay | IC50=155.183 μM | SANGER | |||
| RERF-LC-MS | Growth Inhibition Assay | IC50=154.772 μM | SANGER | |||
| Calu-1 | Growth Inhibition Assay | IC50=154.003 μM | SANGER | |||
| TT | Growth Inhibition Assay | IC50=153.49 μM | SANGER | |||
| MDA-MB-361 | Growth Inhibition Assay | IC50=153.164 μM | SANGER | |||
| AsPC-1 | Growth Inhibition Assay | IC50=152.764 μM | SANGER | |||
| GAK | Growth Inhibition Assay | IC50=151.374 μM | SANGER | |||
| GDM-1 | Growth Inhibition Assay | IC50=151.135 μM | SANGER | |||
| NCI-H748 | Growth Inhibition Assay | IC50=150.209 μM | SANGER | |||
| L-540 | Growth Inhibition Assay | IC50=149.75 μM | SANGER | |||
| Caov-3 | Growth Inhibition Assay | IC50=149.714 μM | SANGER | |||
| TE-9 | Growth Inhibition Assay | IC50=149.443 μM | SANGER | |||
| 639-V | Growth Inhibition Assay | IC50=148.705 μM | SANGER | |||
| LB647-SCLC | Growth Inhibition Assay | IC50=148.562 μM | SANGER | |||
| KYSE-520 | Growth Inhibition Assay | IC50=148.54 μM | SANGER | |||
| GAMG | Growth Inhibition Assay | IC50=148.247 μM | SANGER | |||
| GCIY | Growth Inhibition Assay | IC50=147.797 μM | SANGER | |||
| DMS-114 | Growth Inhibition Assay | IC50=147.441 μM | SANGER | |||
| BB49-HNC | Growth Inhibition Assay | IC50=147.404 μM | SANGER | |||
| SW13 | Growth Inhibition Assay | IC50=147.253 μM | SANGER | |||
| LK-2 | Growth Inhibition Assay | IC50=147.143 μM | SANGER | |||
| OMC-1 | Growth Inhibition Assay | IC50=145.967 μM | SANGER | |||
| G-401 | Growth Inhibition Assay | IC50=145.835 μM | SANGER | |||
| HTC-C3 | Growth Inhibition Assay | IC50=145.075 μM | SANGER | |||
| SCC-25 | Growth Inhibition Assay | IC50=144.005 μM | SANGER | |||
| NCI-H810 | Growth Inhibition Assay | IC50=143.687 μM | SANGER | |||
| CCF-STTG1 | Growth Inhibition Assay | IC50=143.334 μM | SANGER | |||
| YKG-1 | Growth Inhibition Assay | IC50=142.889 μM | SANGER | |||
| OE33 | Growth Inhibition Assay | IC50=141.798 μM | SANGER | |||
| OCI-AML2 | Growth Inhibition Assay | IC50=141.507 μM | SANGER | |||
| Detroit562 | Growth Inhibition Assay | IC50=141.466 μM | SANGER | |||
| OVCAR-4 | Growth Inhibition Assay | IC50=140.749 μM | SANGER | |||
| BHT-101 | Growth Inhibition Assay | IC50=140.208 μM | SANGER | |||
| Ca-Ski | Growth Inhibition Assay | IC50=138.831 μM | SANGER | |||
| OAW-42 | Growth Inhibition Assay | IC50=137.856 μM | SANGER | |||
| G-402 | Growth Inhibition Assay | IC50=135.295 μM | SANGER | |||
| NOS-1 | Growth Inhibition Assay | IC50=134.765 μM | SANGER | |||
| NCI-H1437 | Growth Inhibition Assay | IC50=134.59 μM | SANGER | |||
| JEG-3 | Growth Inhibition Assay | IC50=134.359 μM | SANGER | |||
| CP50-MEL-B | Growth Inhibition Assay | IC50=134.2 μM | SANGER | |||
| BB30-HNC | Growth Inhibition Assay | IC50=134.116 μM | SANGER | |||
| Capan-2 | Growth Inhibition Assay | IC50=133.975 μM | SANGER | |||
| CTB-1 | Growth Inhibition Assay | IC50=132.897 μM | SANGER | |||
| HT-1080 | Growth Inhibition Assay | IC50=132.204 μM | SANGER | |||
| MFH-ino | Growth Inhibition Assay | IC50=131.114 μM | SANGER | |||
| VMRC-RCZ | Growth Inhibition Assay | IC50=130.375 μM | SANGER | |||
| LU-99A | Growth Inhibition Assay | IC50=129.931 μM | SANGER | |||
| HEC-1 | Growth Inhibition Assay | IC50=129.198 μM | SANGER | |||
| EB-3 | Growth Inhibition Assay | IC50=127.013 μM | SANGER | |||
| UMC-11 | Growth Inhibition Assay | IC50=126.038 μM | SANGER | |||
| NCI-H2087 | Growth Inhibition Assay | IC50=124.283 μM | SANGER | |||
| SW1463 | Growth Inhibition Assay | IC50=124.077 μM | SANGER | |||
| OPM-2 | Growth Inhibition Assay | IC50=123.176 μM | SANGER | |||
| HT-29 | Growth Inhibition Assay | IC50=122.778 μM | SANGER | |||
| HAL-01 | Growth Inhibition Assay | IC50=122.499 μM | SANGER | |||
| KYSE-140 | Growth Inhibition Assay | IC50=122.141 μM | SANGER | |||
| HuP-T4 | Growth Inhibition Assay | IC50=121.55 μM | SANGER | |||
| NCI-N87 | Growth Inhibition Assay | IC50=121.044 μM | SANGER | |||
| OVCAR-3 | Growth Inhibition Assay | IC50=119.915 μM | SANGER | |||
| KNS-81-FD | Growth Inhibition Assay | IC50=119.417 μM | SANGER | |||
| SUP-T1 | Growth Inhibition Assay | IC50=118.956 μM | SANGER | |||
| SKG-IIIa | Growth Inhibition Assay | IC50=118.116 μM | SANGER | |||
| RKO | Growth Inhibition Assay | IC50=117.898 μM | SANGER | |||
| ATN-1 | Growth Inhibition Assay | IC50=117.828 μM | SANGER | |||
| OVCAR-8 | Growth Inhibition Assay | IC50=117.668 μM | SANGER | |||
| HCC1599 | Growth Inhibition Assay | IC50=117.156 μM | SANGER | |||
| Calu-6 | Growth Inhibition Assay | IC50=116.798 μM | SANGER | |||
| DMS-273 | Growth Inhibition Assay | IC50=115.617 μM | SANGER | |||
| 786-0 | Growth Inhibition Assay | IC50=113.453 μM | SANGER | |||
| SIG-M5 | Growth Inhibition Assay | IC50=113.125 μM | SANGER | |||
| NEC8 | Growth Inhibition Assay | IC50=112.624 μM | SANGER | |||
| MDA-MB-157 | Growth Inhibition Assay | IC50=112.49 μM | SANGER | |||
| U031 | Growth Inhibition Assay | IC50=112.065 μM | SANGER | |||
| WSU-NHL | Growth Inhibition Assay | IC50=112.01 μM | SANGER | |||
| HCT-15 | Growth Inhibition Assay | IC50=110.661 μM | SANGER | |||
| A253 | Growth Inhibition Assay | IC50=110.479 μM | SANGER | |||
| DU-145 | Growth Inhibition Assay | IC50=109.945 μM | SANGER | |||
| D-263MG | Growth Inhibition Assay | IC50=108.961 μM | SANGER | |||
| FTC-133 | Growth Inhibition Assay | IC50=108.488 μM | SANGER | |||
| HOP-92 | Growth Inhibition Assay | IC50=108.475 μM | SANGER | |||
| NCI-H23 | Growth Inhibition Assay | IC50=108.357 μM | SANGER | |||
| PANC-08-13 | Growth Inhibition Assay | IC50=106.85 μM | SANGER | |||
| RPMI-8226 | Growth Inhibition Assay | IC50=106.678 μM | SANGER | |||
| SNU-C2B | Growth Inhibition Assay | IC50=105.023 μM | SANGER | |||
| UM-UC-3 | Growth Inhibition Assay | IC50=103.842 μM | SANGER | |||
| MKN45 | Growth Inhibition Assay | IC50=103.584 μM | SANGER | |||
| KE-37 | Growth Inhibition Assay | IC50=102.95 μM | SANGER | |||
| SK-MES-1 | Growth Inhibition Assay | IC50=101.762 μM | SANGER | |||
| TE-11 | Growth Inhibition Assay | IC50=101.724 μM | SANGER | |||
| Saos-2 | Growth Inhibition Assay | IC50=101.437 μM | SANGER | |||
| LU-65 | Growth Inhibition Assay | IC50=101.081 μM | SANGER | |||
| Mewo | Growth Inhibition Assay | IC50=99.5603 μM | SANGER | |||
| IGR-1 | Growth Inhibition Assay | IC50=99.3119 μM | SANGER | |||
| NCI-H292 | Growth Inhibition Assay | IC50=99.3004 μM | SANGER | |||
| PC-3 | Growth Inhibition Assay | IC50=99.0564 μM | SANGER | |||
| U-698-M | Growth Inhibition Assay | IC50=98.9489 μM | SANGER | |||
| MKN7 | Growth Inhibition Assay | IC50=97.7439 μM | SANGER | |||
| OS-RC-2 | Growth Inhibition Assay | IC50=97.461 μM | SANGER | |||
| Caov-4 | Growth Inhibition Assay | IC50=95.685 μM | SANGER | |||
| A2058 | Growth Inhibition Assay | IC50=95.4617 μM | SANGER | |||
| RMG-I | Growth Inhibition Assay | IC50=95.1072 μM | SANGER | |||
| MEL-JUSO | Growth Inhibition Assay | IC50=94.954 μM | SANGER | |||
| MZ1-PC | Growth Inhibition Assay | IC50=94.5984 μM | SANGER | |||
| J82 | Growth Inhibition Assay | IC50=93.7811 μM | SANGER | |||
| 23132-87 | Growth Inhibition Assay | IC50=93.168 μM | SANGER | |||
| CAL-33 | Growth Inhibition Assay | IC50=93.0601 μM | SANGER | |||
| BOKU | Growth Inhibition Assay | IC50=92.6957 μM | SANGER | |||
| MZ7-mel | Growth Inhibition Assay | IC50=91.2676 μM | SANGER | |||
| C-33-A | Growth Inhibition Assay | IC50=90.54 μM | SANGER | |||
| NCI-H650 | Growth Inhibition Assay | IC50=89.8724 μM | SANGER | |||
| TE-12 | Growth Inhibition Assay | IC50=89.3741 μM | SANGER | |||
| MDA-MB-175-VII | Growth Inhibition Assay | IC50=85.7426 μM | SANGER | |||
| SBC-1 | Growth Inhibition Assay | IC50=84.0208 μM | SANGER | |||
| BFTC-905 | Growth Inhibition Assay | IC50=84.0172 μM | SANGER | |||
| SW1573 | Growth Inhibition Assay | IC50=82.5893 μM | SANGER | |||
| KMOE-2 | Growth Inhibition Assay | IC50=81.8531 μM | SANGER | |||
| NCI-H1417 | Growth Inhibition Assay | IC50=81.4368 μM | SANGER | |||
| NCI-H661 | Growth Inhibition Assay | IC50=81.344 μM | SANGER | |||
| NB12 | Growth Inhibition Assay | IC50=80.9369 μM | SANGER | |||
| KYSE-410 | Growth Inhibition Assay | IC50=80.2857 μM | SANGER | |||
| HCE-T | Growth Inhibition Assay | IC50=80.1588 μM | SANGER | |||
| SF295 | Growth Inhibition Assay | IC50=79.9553 μM | SANGER | |||
| KP-N-YS | Growth Inhibition Assay | IC50=78.5878 μM | SANGER | |||
| PANC-03-27 | Growth Inhibition Assay | IC50=78.435 μM | SANGER | |||
| NB6 | Growth Inhibition Assay | IC50=78.1299 μM | SANGER | |||
| HSC-3 | Growth Inhibition Assay | IC50=77.0282 μM | SANGER | |||
| 8-MG-BA | Growth Inhibition Assay | IC50=76.1096 μM | SANGER | |||
| SK-MEL-1 | Growth Inhibition Assay | IC50=75.8019 μM | SANGER | |||
| LS-411N | Growth Inhibition Assay | IC50=74.2528 μM | SANGER | |||
| RT4 | Growth Inhibition Assay | IC50=73.5742 μM | SANGER | |||
| KALS-1 | Growth Inhibition Assay | IC50=73.5475 μM | SANGER | |||
| EW-16 | Growth Inhibition Assay | IC50=71.7415 μM | SANGER | |||
| HCC1806 | Growth Inhibition Assay | IC50=71.4587 μM | SANGER | |||
| Hs-578-T | Growth Inhibition Assay | IC50=70.3 μM | SANGER | |||
| NCI-H2342 | Growth Inhibition Assay | IC50=69.3961 μM | SANGER | |||
| C-4-II | Growth Inhibition Assay | IC50=69.3607 μM | SANGER | |||
| NCI-H1703 | Growth Inhibition Assay | IC50=68.1171 μM | SANGER | |||
| HuCCT1 | Growth Inhibition Assay | IC50=67.0374 μM | SANGER | |||
| MFE-296 | Growth Inhibition Assay | IC50=64.4473 μM | SANGER | |||
| AN3-CA | Growth Inhibition Assay | IC50=64.3035 μM | SANGER | |||
| SCH | Growth Inhibition Assay | IC50=64.2351 μM | SANGER | |||
| T-24 | Growth Inhibition Assay | IC50=62.7126 μM | SANGER | |||
| K-562 | Growth Inhibition Assay | IC50=62.2452 μM | SANGER | |||
| NCI-H1734 | Growth Inhibition Assay | IC50=61.9663 μM | SANGER | |||
| NMC-G1 | Growth Inhibition Assay | IC50=61.9242 μM | SANGER | |||
| EKVX | Growth Inhibition Assay | IC50=61.4958 μM | SANGER | |||
| SiHa | Growth Inhibition Assay | IC50=59.6487 μM | SANGER | |||
| LU-134-A | Growth Inhibition Assay | IC50=59.6016 μM | SANGER | |||
| ACN | Growth Inhibition Assay | IC50=59.1587 μM | SANGER | |||
| HT-1376 | Growth Inhibition Assay | IC50=59.0921 μM | SANGER | |||
| LCLC-97TM1 | Growth Inhibition Assay | IC50=59.0553 μM | SANGER | |||
| BV-173 | Growth Inhibition Assay | IC50=57.5142 μM | SANGER | |||
| RPMI-8866 | Growth Inhibition Assay | IC50=57.2412 μM | SANGER | |||
| DoTc2-4510 | Growth Inhibition Assay | IC50=56.6057 μM | SANGER | |||
| NCI-H2405 | Growth Inhibition Assay | IC50=56.1693 μM | SANGER | |||
| RCM-1 | Growth Inhibition Assay | IC50=56.0078 μM | SANGER | |||
| DV-90 | Growth Inhibition Assay | IC50=55.8917 μM | SANGER | |||
| ES6 | Growth Inhibition Assay | IC50=55.8899 μM | SANGER | |||
| LXF-289 | Growth Inhibition Assay | IC50=55.5066 μM | SANGER | |||
| A375 | Growth Inhibition Assay | IC50=54.8059 μM | SANGER | |||
| MC-IXC | Growth Inhibition Assay | IC50=54.4364 μM | SANGER | |||
| 697 | Growth Inhibition Assay | IC50=54.1555 μM | SANGER | |||
| KGN | Growth Inhibition Assay | IC50=53.959 μM | SANGER | |||
| MG-63 | Growth Inhibition Assay | IC50=53.9381 μM | SANGER | |||
| COLO-824 | Growth Inhibition Assay | IC50=53.6976 μM | SANGER | |||
| CHP-126 | Growth Inhibition Assay | IC50=53.5169 μM | SANGER | |||
| CESS | Growth Inhibition Assay | IC50=52.145 μM | SANGER | |||
| DB | Growth Inhibition Assay | IC50=51.9855 μM | SANGER | |||
| SW837 | Growth Inhibition Assay | IC50=51.9785 μM | SANGER | |||
| SK-N-AS | Growth Inhibition Assay | IC50=51.8281 μM | SANGER | |||
| MCF7 | Growth Inhibition Assay | IC50=51.7554 μM | SANGER | |||
| NCI-H1650 | Growth Inhibition Assay | IC50=51.5849 μM | SANGER | |||
| NCI-H1793 | Growth Inhibition Assay | IC50=51.231 μM | SANGER | |||
| KS-1 | Growth Inhibition Assay | IC50=50.2225 μM | SANGER | |||
| LC4-1 | Growth Inhibition Assay | IC50=49.8912 μM | SANGER | |||
| SNU-C1 | Growth Inhibition Assay | IC50=49.3207 μM | SANGER | |||
| COR-L105 | Growth Inhibition Assay | IC50=48.973 μM | SANGER | |||
| SNB75 | Growth Inhibition Assay | IC50=48.4118 μM | SANGER | |||
| HCC2998 | Growth Inhibition Assay | IC50=48.2982 μM | SANGER | |||
| NCI-H747 | Growth Inhibition Assay | IC50=47.7264 μM | SANGER | |||
| D-542MG | Growth Inhibition Assay | IC50=47.4589 μM | SANGER | |||
| UACC-62 | Growth Inhibition Assay | IC50=46.8608 μM | SANGER | |||
| no-11 | Growth Inhibition Assay | IC50=46.2031 μM | SANGER | |||
| Ramos-2G6-4C10 | Growth Inhibition Assay | IC50=46.0755 μM | SANGER | |||
| COLO-800 | Growth Inhibition Assay | IC50=45.5898 μM | SANGER | |||
| H4 | Growth Inhibition Assay | IC50=45.3296 μM | SANGER | |||
| KYSE-270 | Growth Inhibition Assay | IC50=45.0437 μM | SANGER | |||
| ESS-1 | Growth Inhibition Assay | IC50=44.2062 μM | SANGER | |||
| SR | Growth Inhibition Assay | IC50=43.8673 μM | SANGER | |||
| TE-8 | Growth Inhibition Assay | IC50=43.7895 μM | SANGER | |||
| LB831-BLC | Growth Inhibition Assay | IC50=43.697 μM | SANGER | |||
| HCT-116 | Growth Inhibition Assay | IC50=43.2099 μM | SANGER | |||
| IST-MEL1 | Growth Inhibition Assay | IC50=42.6938 μM | SANGER | |||
| NY | Growth Inhibition Assay | IC50=42.1801 μM | SANGER | |||
| MPP-89 | Growth Inhibition Assay | IC50=41.9211 μM | SANGER | |||
| SK-CO-1 | Growth Inhibition Assay | IC50=41.7755 μM | SANGER | |||
| BE-13 | Growth Inhibition Assay | IC50=41.4473 μM | SANGER | |||
| HCC2218 | Growth Inhibition Assay | IC50=40.5849 μM | SANGER | |||
| KM12 | Growth Inhibition Assay | IC50=38.5998 μM | SANGER | |||
| CMK | Growth Inhibition Assay | IC50=38.4162 μM | SANGER | |||
| HT-144 | Growth Inhibition Assay | IC50=38.2575 μM | SANGER | |||
| no-10 | Growth Inhibition Assay | IC50=38.1294 μM | SANGER | |||
| MKN1 | Growth Inhibition Assay | IC50=36.8444 μM | SANGER | |||
| SK-UT-1 | Growth Inhibition Assay | IC50=36.3331 μM | SANGER | |||
| 8505C | Growth Inhibition Assay | IC50=35.9294 μM | SANGER | |||
| EFE-184 | Growth Inhibition Assay | IC50=35.917 μM | SANGER | |||
| IGROV-1 | Growth Inhibition Assay | IC50=35.3416 μM | SANGER | |||
| TK10 | Growth Inhibition Assay | IC50=35.1322 μM | SANGER | |||
| PFSK-1 | Growth Inhibition Assay | IC50=34.9241 μM | SANGER | |||
| KU-19-19 | Growth Inhibition Assay | IC50=33.7534 μM | SANGER | |||
| RVH-421 | Growth Inhibition Assay | IC50=33.168 μM | SANGER | |||
| NCI-H1648 | Growth Inhibition Assay | IC50=32.8467 μM | SANGER | |||
| HCC1143 | Growth Inhibition Assay | IC50=32.7979 μM | SANGER | |||
| SW948 | Growth Inhibition Assay | IC50=32.7548 μM | SANGER | |||
| KNS-42 | Growth Inhibition Assay | IC50=31.8662 μM | SANGER | |||
| CAS-1 | Growth Inhibition Assay | IC50=31.7665 μM | SANGER | |||
| LB373-MEL-D | Growth Inhibition Assay | IC50=31.0047 μM | SANGER | |||
| SK-MM-2 | Growth Inhibition Assay | IC50=30.9792 μM | SANGER | |||
| NALM-6 | Growth Inhibition Assay | IC50=30.4965 μM | SANGER | |||
| CAKI-1 | Growth Inhibition Assay | IC50=30.2656 μM | SANGER | |||
| LCLC-103H | Growth Inhibition Assay | IC50=30.2059 μM | SANGER | |||
| IST-MES1 | Growth Inhibition Assay | IC50=29.7092 μM | SANGER | |||
| A204 | Growth Inhibition Assay | IC50=29.6828 μM | SANGER | |||
| JiyoyeP-2003 | Growth Inhibition Assay | IC50=29.6719 μM | SANGER | |||
| SW872 | Growth Inhibition Assay | IC50=28.5326 μM | SANGER | |||
| C32 | Growth Inhibition Assay | IC50=28.0826 μM | SANGER | |||
| OAW-28 | Growth Inhibition Assay | IC50=28.0052 μM | SANGER | |||
| SU-DHL-1 | Growth Inhibition Assay | IC50=27.9474 μM | SANGER | |||
| MC-CAR | Growth Inhibition Assay | IC50=27.7907 μM | SANGER | |||
| ES8 | Growth Inhibition Assay | IC50=27.7814 μM | SANGER | |||
| COLO-792 | Growth Inhibition Assay | IC50=27.7728 μM | SANGER | |||
| NCI-H2170 | Growth Inhibition Assay | IC50=27.5282 μM | SANGER | |||
| KLE | Growth Inhibition Assay | IC50=27.2565 μM | SANGER | |||
| SNG-M | Growth Inhibition Assay | IC50=27.2271 μM | SANGER | |||
| NCI-H1770 | Growth Inhibition Assay | IC50=26.8986 μM | SANGER | |||
| RO82-W-1 | Growth Inhibition Assay | IC50=26.7377 μM | SANGER | |||
| KASUMI-1 | Growth Inhibition Assay | IC50=24.9328 μM | SANGER | |||
| LU-139 | Growth Inhibition Assay | IC50=24.835 μM | SANGER | |||
| NCI-H1963 | Growth Inhibition Assay | IC50=24.2839 μM | SANGER | |||
| HMV-II | Growth Inhibition Assay | IC50=24.2163 μM | SANGER | |||
| PANC-10-05 | Growth Inhibition Assay | IC50=24.0898 μM | SANGER | |||
| K5 | Growth Inhibition Assay | IC50=23.5486 μM | SANGER | |||
| REH | Growth Inhibition Assay | IC50=23.1699 μM | SANGER | |||
| M14 | Growth Inhibition Assay | IC50=23.1132 μM | SANGER | |||
| GI-1 | Growth Inhibition Assay | IC50=22.7841 μM | SANGER | |||
| KURAMOCHI | Growth Inhibition Assay | IC50=22.7069 μM | SANGER | |||
| LS-513 | Growth Inhibition Assay | IC50=22.6192 μM | SANGER | |||
| RXF393 | Growth Inhibition Assay | IC50=22.6068 μM | SANGER | |||
| SW684 | Growth Inhibition Assay | IC50=22.2858 μM | SANGER | |||
| KARPAS-299 | Growth Inhibition Assay | IC50=22.1954 μM | SANGER | |||
| TGBC24TKB | Growth Inhibition Assay | IC50=21.8957 μM | SANGER | |||
| EoL-1- | Growth Inhibition Assay | IC50=21.8385 μM | SANGER | |||
| BPH-1 | Growth Inhibition Assay | IC50=21.7035 μM | SANGER | |||
| SK-MEL-28 | Growth Inhibition Assay | IC50=21.6863 μM | SANGER | |||
| KYSE-450 | Growth Inhibition Assay | IC50=21.5354 μM | SANGER | |||
| HCC1187 | Growth Inhibition Assay | IC50=20.3589 μM | SANGER | |||
| NCI-H727 | Growth Inhibition Assay | IC50=20.2169 μM | SANGER | |||
| Becker | Growth Inhibition Assay | IC50=20.1777 μM | SANGER | |||
| MHH-NB-11 | Growth Inhibition Assay | IC50=19.5342 μM | SANGER | |||
| A101D | Growth Inhibition Assay | IC50=19.243 μM | SANGER | |||
| SNU-449 | Growth Inhibition Assay | IC50=18.715 μM | SANGER | |||
| EM-2 | Growth Inhibition Assay | IC50=18.6248 μM | SANGER | |||
| 8305C | Growth Inhibition Assay | IC50=18.5305 μM | SANGER | |||
| SIMA | Growth Inhibition Assay | IC50=17.9118 μM | SANGER | |||
| Daudi | Growth Inhibition Assay | IC50=17.6149 μM | SANGER | |||
| CCRF-CEM | Growth Inhibition Assay | IC50=16.6702 μM | SANGER | |||
| NH-12 | Growth Inhibition Assay | IC50=16.0097 μM | SANGER | |||
| LoVo | Growth Inhibition Assay | IC50=15.2659 μM | SANGER | |||
| ST486 | Growth Inhibition Assay | IC50=14.2251 μM | SANGER | |||
| NB1 | Growth Inhibition Assay | IC50=14.1935 μM | SANGER | |||
| SF539 | Growth Inhibition Assay | IC50=14.0116 μM | SANGER | |||
| SW48 | Growth Inhibition Assay | IC50=13.7265 μM | SANGER | |||
| MEL-HO | Growth Inhibition Assay | IC50=13.8829 μM | SANGER | |||
| KU812 | Growth Inhibition Assay | IC50=13.5175 μM | SANGER | |||
| T98G | Growth Inhibition Assay | IC50=13.4717 μM | SANGER | |||
| DSH1 | Growth Inhibition Assay | IC50=13.3879 μM | SANGER | |||
| NCI-H82 | Growth Inhibition Assay | IC50=13.2909 μM | SANGER | |||
| C8166 | Growth Inhibition Assay | IC50=13.2318 μM | SANGER | |||
| PC-14 | Growth Inhibition Assay | IC50=12.324 μM | SANGER | |||
| PA-1 | Growth Inhibition Assay | IC50=11.93 μM | SANGER | |||
| HEL | Growth Inhibition Assay | IC50=11.6579 μM | SANGER | |||
| NCI-H526 | Growth Inhibition Assay | IC50=11.1365 μM | SANGER | |||
| SNU-387 | Growth Inhibition Assay | IC50=10.7235 μM | SANGER | |||
| HT-3 | Growth Inhibition Assay | IC50=10.6179 μM | SANGER | |||
| M059J | Growth Inhibition Assay | IC50=10.6102 μM | SANGER | |||
| MZ2-MEL | Growth Inhibition Assay | IC50=10.4677 μM | SANGER | |||
| NCI-H28 | Growth Inhibition Assay | IC50=10.4413 μM | SANGER | |||
| LB1047-RCC | Growth Inhibition Assay | IC50=9.99798 μM | SANGER | |||
| U-118-MG | Growth Inhibition Assay | IC50=9.94444 μM | SANGER | |||
| DBTRG-05MG | Growth Inhibition Assay | IC50=9.7777 μM | SANGER | |||
| EFM-19 | Growth Inhibition Assay | IC50=9.75566 μM | SANGER | |||
| SK-N-DZ | Growth Inhibition Assay | IC50=9.65742 μM | SANGER | |||
| LB996-RCC | Growth Inhibition Assay | IC50=9.52397 μM | SANGER | |||
| U-2-OS | Growth Inhibition Assay | IC50=9.45006 μM | SANGER | |||
| CTV-1 | Growth Inhibition Assay | IC50=9.43894 μM | SANGER | |||
| COLO-205 | Growth Inhibition Assay | IC50=9.43821 μM | SANGER | |||
| NCI-H64 | Growth Inhibition Assay | IC50=9.42255 μM | SANGER | |||
| NCI-H1755 | Growth Inhibition Assay | IC50=9.39986 μM | SANGER | |||
| ACHN | Growth Inhibition Assay | IC50=9.30546 μM | SANGER | |||
| MEG-01 | Growth Inhibition Assay | IC50=8.99043 μM | SANGER | |||
| MKN28 | Growth Inhibition Assay | IC50=8.88516 μM | SANGER | |||
| CAL-72 | Growth Inhibition Assay | IC50=8.76113 μM | SANGER | |||
| MV-4-11 | Growth Inhibition Assay | IC50=8.54304 μM | SANGER | |||
| SW1417 | Growth Inhibition Assay | IC50=7.64584 μM | SANGER | |||
| G-361 | Growth Inhibition Assay | IC50=7.46561 μM | SANGER | |||
| MC116 | Growth Inhibition Assay | IC50=7.28691 μM | SANGER | |||
| RT-112 | Growth Inhibition Assay | IC50=7.16456 μM | SANGER | |||
| CAL-85-1 | Growth Inhibition Assay | IC50=6.60936 μM | SANGER | |||
| BB65-RCC | Growth Inhibition Assay | IC50=6.53195 μM | SANGER | |||
| CGTH-W-1 | Growth Inhibition Assay | IC50=6.417 μM | SANGER | |||
| WM-115 | Growth Inhibition Assay | IC50=5.81509 μM | SANGER | |||
| SK-MEL-2 | Growth Inhibition Assay | IC50=6.38344 μM | SANGER | |||
| BL-41 | Growth Inhibition Assay | IC50=5.81266 μM | SANGER | |||
| LB2518-MEL | Growth Inhibition Assay | IC50=5.72627 μM | SANGER | |||
| D-392MG | Growth Inhibition Assay | IC50=5.49145 μM | SANGER | |||
| NCI-H2452 | Growth Inhibition Assay | IC50=5.45628 μM | SANGER | |||
| SW1710 | Growth Inhibition Assay | IC50=5.42456 μM | SANGER | |||
| SK-OV-3 | Growth Inhibition Assay | IC50=4.9934 μM | SANGER | |||
| ALL-PO | Growth Inhibition Assay | IC50=4.83858 μM | SANGER | |||
| BxPC-3 | Growth Inhibition Assay | IC50=4.70391 μM | SANGER | |||
| RCC10RGB | Growth Inhibition Assay | IC50=4.29479 μM | SANGER | |||
| NKM-1 | Growth Inhibition Assay | IC50=4.13971 μM | SANGER | |||
| MHH-ES-1 | Growth Inhibition Assay | IC50=4.04545 μM | SANGER | |||
| OE19 | Growth Inhibition Assay | IC50=3.96451 μM | SANGER | |||
| TE-15 | Growth Inhibition Assay | IC50=3.95343 μM | SANGER | |||
| MOLT-16 | Growth Inhibition Assay | IC50=3.88556 μM | SANGER | |||
| A172 | Growth Inhibition Assay | IC50=3.75498 μM | SANGER | |||
| HCC1569 | Growth Inhibition Assay | IC50=3.48305 μM | SANGER | |||
| HOP-62 | Growth Inhibition Assay | IC50=3.36054 μM | SANGER | |||
| A704 | Growth Inhibition Assay | IC50=3.0812 μM | SANGER | |||
| SCC-9 | Growth Inhibition Assay | IC50=2.95485 μM | SANGER | |||
| COLO-829 | Growth Inhibition Assay | IC50=2.7654 μM | SANGER | |||
| RPMI-7951 | Growth Inhibition Assay | IC50=2.70193 μM | SANGER | |||
| SK-LMS-1 | Growth Inhibition Assay | IC50=2.64128 μM | SANGER | |||
| SNU-423 | Growth Inhibition Assay | IC50=2.49787 μM | SANGER | |||
| NCI-H2122 | Growth Inhibition Assay | IC50=2.27684 μM | SANGER | |||
| RS4-11 | Growth Inhibition Assay | IC50=1.95415 μM | SANGER | |||
| HCC1395 | Growth Inhibition Assay | IC50=1.78273 μM | SANGER | |||
| MIA-PaCa-2 | Growth Inhibition Assay | IC50=1.75977 μM | SANGER | |||
| KYSE-180 | Growth Inhibition Assay | IC50=1.72788 μM | SANGER | |||
| BCPAP | Growth Inhibition Assay | IC50=1.66898 μM | SANGER | |||
| A498 | Growth Inhibition Assay | IC50=1.30469 μM | SANGER | |||
| TYK-nu | Growth Inhibition Assay | IC50=1.07912 μM | SANGER | |||
| D-247MG | Growth Inhibition Assay | IC50=0.94852 μM | SANGER | |||
| S-117 | Growth Inhibition Assay | IC50=0.92131 μM | SANGER | |||
| D-336MG | Growth Inhibition Assay | IC50=0.8462 μM | SANGER | |||
| LOXIMVI | Growth Inhibition Assay | IC50=0.58832 μM | SANGER | |||
| 769-P | Growth Inhibition Assay | IC50=0.58499 μM | SANGER | |||
| GCT | Growth Inhibition Assay | IC50=0.30672 μM | SANGER | |||
| SW982 | Growth Inhibition Assay | IC50=0.21416 μM | SANGER | |||
| ES-CCE | Function Assay | 5 μM | 3 d | DMSO | Strongly induces the activity of Wnt/beta-catenin pathway | 24779365 |
| ES-D3 | Function Assay | 5 μM | 3 d | DMSO | Strongly induces the activity of Wnt/beta-catenin pathway | 24779365 |
| ES-CCE | Cytotoxic Assay | 10 μM | 3 d | DMSO | Induces moderate cytotoxicity | 24779365 |
| ES-D3 | Cytotoxic Assay | 10 μM | 3 d | DMSO | Induces moderate cytotoxicity with IC50 of 4.9 μM | 24779365 |
| Neural progenitor cells | Function Assay | 3 μM | 10 d | DMSO | Induces generation of neural progenitor cells | 24638034 |
| A549 | Apoptosis Assay | 5 µM | 24 h | DMSO | Increases cell apoptosis | 24367688 |
| H157 | Apoptosis Assay | 5 µM | 24 h | DMSO | Does not significantly affect apoptosis | 24367688 |
| H460 | Apoptosis Assay | 5 µM | 24 h | DMSO | Attenuates cell apoptosis | 24367688 |
| H157 | Growth Inhibition Assay | 5 µM | 24 h | DMSO | Promotes cell proliferation | 24367688 |
| H460 | Growth Inhibition Assay | 5 µM | 24 h | DMSO | Promotes cell proliferation | 24367688 |
| embryonic stem cells (ESC) | Function Assay | 3 µM | 11 d | DMSO | Promotes self-renewal of naïve-type mouse embryonic stem cells (ESC) | 23843312 |
| DN3 | Function Assay | 10 μM | 96 h | DMSO | Increases cell development and proliferation | 23526989 |
| platelets | Function Assay | 10 μM | 15 min | DMSO | Increases thrombin-mediated platelet aggregation and ATP secretion | 23239877 |
| hiPSC-NPs | Growth Inhibition Assay | 3 μM | 72 h | DMSO | Increases cell proliferation | 22642687 |
| INS-1E | Function Assay | 2.5 μM | 24 h | DMSO | Induces a reduction of NP1 overexpression | 22427704 |
| PANC1 | Apoptosis Assay | 5 μM | 72 h | DMSO | Triggers an apoptotic response | 22201186 |
| Endothelial cell(EC) | Function Assay | 3 μM | DMSO | Promotes endothelial cell (EC) differentiation | 22052006 | |
| HMC1.2 | Growth Inhibition Assay | 30 μM | 72 h | DMSO | Reduces cell survival | 22039301 |
| Sca-1+ HPC | Growth Inhibition Assay | 2 μM | 6 d | DMSO | Synergistically promotes Sca-1+ HPC proliferation and colony formation together with EGF | 21624348 |
| Oct4/Sox2/Klf4-infected MEFs | Function Assay | 10 μM | 28 d | DMSO | Promotes MEFs reprogramming Transduced by Oct4, Sox2, and Klf4 | 19839055 |
| H4IIE | Function Assay | 10 μM | 24 h | DMSO | Regulates β-catenin activity and TIRE containing promoter activity | 15350195 |
| insulin receptor–expressing CHO-IR | Function Assay | 500 nM | 30 min | DMSO | Activates Glycogen Synthase (GS) | 12606497 |
| 3T3-L1 | Function Assay | 3 μM | 4 h | DMSO | Inhibits preadipocyte differentiation | 12055200 |
| CHO-IR | Function Assay | 30 min | EC50=0.77 μM | 29016121 | ||
| HepG2 | Function Assay | 3 h | EC50=1.5 μM | 22261023 | ||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 93 mg/mL
(199.85 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 465.34 | Formula | C22H18Cl2N8 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 252917-06-9 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | CT99021 | Smiles | CC1=CN=C(N1)C2=CN=C(N=C2C3=C(C=C(C=C3)Cl)Cl)NCCNC4=NC=C(C=C4)C#N | ||
Mechanism of Action
| Targets/IC50/Ki |
GSK-3β
(Cell-free assay) 6.7 nM
GSK-3α
(Cell-free assay) 10 nM
|
|---|---|
| In vitro |
CHIR-99021 (Laduviglusib) shows greater than 500-fold selectivity for GSK-3 versus its closest homologs CDC2 and ERK2, as well as other protein kinases. Furthermore, it shows only weak binding to a panel of 22 pharmacologically relevant receptors and little inhibitory activity against a panel of 23 nonkinase enzymes. This compound induces the activation of glycogen synthase (GS) in insulin receptor-expressing CHO-IR cells with EC50 of 0.763 μM.
|
| In vivo |
Oral administration of CHIR-99021 (Laduviglusib) at 30 mg/kg enhances glucose metabolism in a rodent model of type 2 diabetes, with a maximal plasma glucose reduction of nearly 150 mg/dl 3-4 hours after administration, while plasma insulin remains at or below control levels. When given orally at 16 or 48 mg/kg 1 hour before oral glucose challenges in ZDF rats, this compound significantly improves glucose tolerance with 14% and 33% reduction in plasma glucose at 16 mg/kg and 48 mg/kg, respectively; the higher dose also reduces hyperglycemia before the oral glucose challenge.
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | Cleaved PARP / CD31 / VE-Cadherin / α-SMA / vimentin CTNNB1 β-catenin |
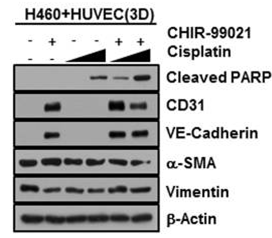
|
30709379 |
| Growth inhibition assay | Cell viability |
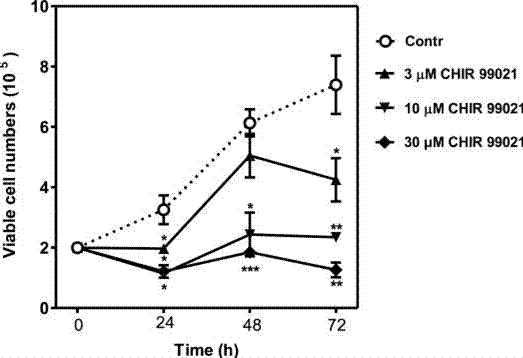
|
22039301 |
| Immunofluorescence | SOX10/EdU Oct3/4 |
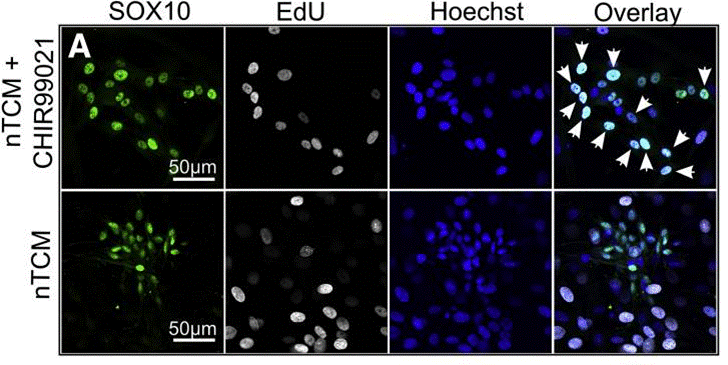
|
28174705 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































