research use only
Rucaparib phosphate PARP inhibitor
Cat.No.S1098
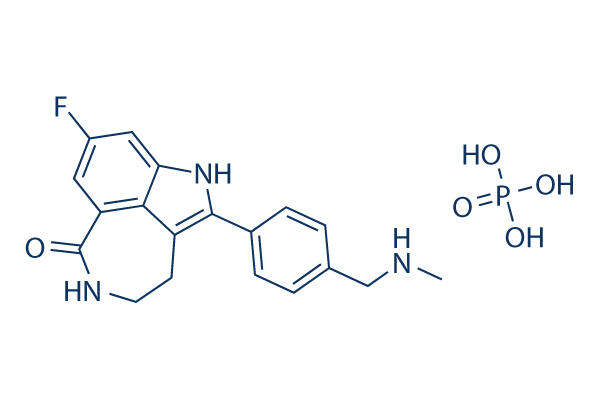
Chemical Structure
Molecular Weight: 421.36
Quality Control
| Related Targets | HDAC ATM/ATR DNA-PK WRN DNA/RNA Synthesis Topoisomerase PPAR Sirtuin Casein Kinase eIF |
|---|---|
| Other PARP Inhibitors | XAV-939 AZD5305 (Saruparib) Veliparib (ABT-888) PJ34 HCl AG-14361 Iniparib (BSI-201) G007-LK Pamiparib UPF 1069 A-966492 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| BT-474 | Function Assay | 0.1/1/500/1000 nM | inhibits PARP activity at starting concerntration of 500 nM | 25128455 | ||
| BT474 | Growth Inhibition Assay | 500 nM | 10–15 d | reduces cell growth in the four lines and significantly | 25128455 | |
| SKBR3 | Growth Inhibition Assay | 500 nM | 10–15 d | reduces cell growth in the four lines and significantly | 25128455 | |
| AU565 | Growth Inhibition Assay | 500 nM | 10–15 d | reduces cell growth in the four lines and significantly | 25128455 | |
| EFM192A | Growth Inhibition Assay | 500 nM | 10–15 d | reduces cell growth in the four lines and significantly | 25128455 | |
| MDA-MB-231 | Function Assay | 10/20/40 μM | 24 h | increases p-AKT levels in a dose-dependent manner | 24420152 | |
| MDA-MB-231 | Cell Viability Assay | 0.1-40 μM | 24 h | IC50 = 17.77 μM | 24420152 | |
| MDA-MB-231 | Apoptosis Assay | 10/20/40 μM | 24 h | induces apoptosis dose dependently | 24420152 | |
| MDA-MB-231 | Growth Inhibition Assay | 10/20/40 μM | 24 h | blocks cell cycle progression in G2/M phase | 24420152 | |
| H460 | Growth Inhibition Assay | 400 nM | 24 h | increases cellular radiosensitivity | 24411611 | |
| A549 | Growth Inhibition Assay | 400 nM | 24 h | increases cellular radiosensitivity | 24411611 | |
| DT40 | Growth Inhibition Assay | IC50=21 nM | 24356813 | |||
| DU145 | Growth Inhibition Assay | IC50=18 nM | 24356813 | |||
| COLO704 | Growth Inhibition Assay | IC50=2.52 ± 0.67 μM | 23729402 | |||
| OVMANAb | Growth Inhibition Assay | IC50=2.58 ± 0.38 μM | 23729402 | |||
| OV177 | Growth Inhibition Assay | IC50=2.78 ± 0.71 μM | 23729402 | |||
| OAW28 | Growth Inhibition Assay | IC50=3.61 ± 0.28 μM | 23729402 | |||
| OVSAHO | Growth Inhibition Assay | IC50=3.64 ± 0.33 μM | 23729402 | |||
| OVKATE | Growth Inhibition Assay | IC50=3.64 ± 1.79 μM | 23729402 | |||
| OVCAR3 | Growth Inhibition Assay | IC50=3.74 ± 0.40 μM | 23729402 | |||
| PEO14 | Growth Inhibition Assay | IC50=3.84 ± 0.76 μM | 23729402 | |||
| A2780 | Growth Inhibition Assay | IC50=3.94 ± 0.25 μM | 23729402 | |||
| OVTOKO | Growth Inhibition Assay | IC50=4.14 ± 1.53 μM | 23729402 | |||
| KURAMOCHIb | Growth Inhibition Assay | IC50=4.34 ± 0.29 μM | 23729402 | |||
| TOV21G | Growth Inhibition Assay | IC50=5.07 ± 1.30 μM | 23729402 | |||
| OVISE | Growth Inhibition Assay | IC50=5.68 ± 0.23 μM | 23729402 | |||
| KK | Growth Inhibition Assay | IC50=6.15 ± 1.42 μM | 23729402 | |||
| RMUGS | Growth Inhibition Assay | IC50=7.03 ± 1.83 μM | 23729402 | |||
| PEO6 | Growth Inhibition Assay | IC50=7.06 ± 0.74 μM | 23729402 | |||
| OVCA429 | Growth Inhibition Assay | IC50=8.29 ± 1.64 μM | 23729402 | |||
| OV167 | Growth Inhibition Assay | IC50=8.33 ± 1.18 μM | 23729402 | |||
| RMG1 | Growth Inhibition Assay | IC50=9.32 ± 2.36 μM | 23729402 | |||
| OVCAR5 | Growth Inhibition Assay | IC50=9.50 ± 2.59 μM | 23729402 | |||
| EFO21 | Growth Inhibition Assay | IC50=9.92 ± 1.87 μM | 23729402 | |||
| ES2 | Growth Inhibition Assay | IC50=10.12 ± 1.23 μM | 23729402 | |||
| Tyk-nu | Growth Inhibition Assay | IC50=10.20 ± 1.12 μM | 23729402 | |||
| CAOV3 | Growth Inhibition Assay | IC50=10.37 ± 0.87 μM | 23729402 | |||
| OV207 | Growth Inhibition Assay | IC50=12.27 ± 0.32 μM | 23729402 | |||
| HEY | Growth Inhibition Assay | IC50=13.01 ± 0.75 μM | 23729402 | |||
| DOV13 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| EFO27 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| HEY C2 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| KOC-7cc | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| MCASb | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| OAW42 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| OV2008 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| OV90 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| OVCA420b | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| OVCA432 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| PEA2 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| SKOV3 | Growth Inhibition Assay | IC50>15 μM | 23729402 | |||
| TOV112D | Growth Inhibition Assay | 0-3 μM | IC50>15 μM | 23729402 | ||
| C4-2 | Growth Inhibition Assay | 0-3 μM | 14 d | DMSO | decreases colony number dose dependently | 23565244 |
| PC3 | Growth Inhibition Assay | 0-3 μM | 14 d | DMSO | decreases colony number dose dependently | 23565244 |
| DU145 | Growth Inhibition Assay | 0-3 μM | 14 d | DMSO | decreases colony number dose dependently | 23565244 |
| VCaP | Growth Inhibition Assay | 0-3 μM | 14 d | DMSO | decreases colony number dose dependently | 23565244 |
| LNCaP | Growth Inhibition Assay | 0-3 μM | 14 d | DMSO | decreases colony number dose dependently | 23565244 |
| MDA-MB-468 | Cell Viability Assay | IC50=9.7 μM | 22678161 | |||
| MDA-MB-231 | Cell Viability Assay | IC50=13 μM | 22678161 | |||
| Cal-51 | Cell Viability Assay | IC50=8.6 μM | 22678161 | |||
| LoVo | Function assay | 30 mins | Inhibition of PARP in human LoVo cells assessed as inhibition of poly(ADP)-ribose polymerization for 30 mins by fluorescence assay, EC50 = 0.00469 μM. | 26652717 | ||
| MX1 | Cytotoxicity assay | Cytotoxicity against BRCA1-deficient human MX1 cells, EC50 = 0.0053 μM. | 26652717 | |||
| Rosetta2 (DE3) | Function assay | Inhibition of human N-terminal 6xhis-tagged ARTD6 (873 to 1161) expressed in Escherichia coli Rosetta2 (DE3) cells using NAD+ as substrate by fluorescence assay, IC50 = 0.014 μM. | 24900770 | |||
| Rosetta2 (DE3) | Function assay | Inhibition of human 6xhis-tagged ARTD5 (1030 to 1317) expressed in Escherichia coli Rosetta2 (DE3) cells using NAD+ as substrate by fluorescence assay, IC50 = 0.025 μM. | 24900770 | |||
| LoVo | Cytotoxicity assay | 0.4 uM | 5 days | Potentiation of temozolomide-induced cytotoxicity in human LoVo cells assessed as temozolomide GI50 at 0.4 uM after 5 days by Celltiter-Glo assay, GI50 = 0.144 μM. | 26652717 | |
| Capan1 | Cytotoxicity assay | Cytotoxicity against BRCA2-deficient human Capan1 cells, EC50 = 0.609 μM. | 26652717 | |||
| MDA-MB-436 | Anticancer assay | 96 hrs | Anticancer activity against human BRCA1-deficient MDA-MB-436 cells after 96 hrs by MTT assay, CC50 = 3 μM. | 26342868 | ||
| OVCAR3 | Antiproliferative assay | 24 hrs | Antiproliferative activity against human OVCAR3 cells after 24 hrs by MTT assay, IC50 = 3.31 μM. | 29456106 | ||
| MRC5 | Cytotoxicity assay | Cytotoxicity against human MRC5 cells, EC50 = 8.53 μM. | 26652717 | |||
| MCF7 | Anticancer assay | 96 hrs | Anticancer activity against human MCF7 cells after 96 hrs by MTT assay, CC50 = 19.47 μM. | 26342868 | ||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for A673 cells) | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh41 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-MC cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for TC32 cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 85 mg/mL
(201.72 mM)
Water : 7 mg/mL Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 421.36 | Formula | C19H18FN3O.H3PO4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 459868-92-9 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | AG-014699 phosphate, PF-01367338 phosphate | Smiles | CNCC1=CC=C(C=C1)C2=C3CCNC(=O)C4=C3C(=CC(=C4)F)N2.OP(=O)(O)O | ||
Mechanism of Action
| Features |
The first PARP inhibitor used in clinical trials combined with temozolomide.
|
|---|---|
| Targets/IC50/Ki |
PARP
(Cell-free assay) 1.4 nM(Ki)
|
| In vitro |
Rucaparib is a potent inhibitor of purified full-length human PARP-1 and shows higher inhibition of cellular PARP in LoVo and SW620 cells. Besides, Rucaparib binds detectably to eight other PARP domains, including PARP2, 3, 4, 10, 15, 16, TNKS1 and TNKS2. The radio-sensitization by Rucaparib is due to downstream inhibition of activation of NF-κB, and is independent of SSB repair inhibition. Rucaparib could target NF-κB activated by DNA damage and overcome toxicity observed with classical NF-κB inhibitors without compromising other vital inflammatory functions. Rucaparib inhibits PARP-1 activity by 97.1% at a concentration of 1 μM in permeabilised D283Med cells.
|
| Kinase Assay |
Ki Determination
|
|
Inhibition of human full-length recombinant PARP-1 by [32P]NAD+ incorporation is measured. The [32P]ADP-ribose incorporated into acid insoluble material is quantified using a PhosphorImager. Ki is calculated by nonlinear regression analysis.
|
|
| In vivo |
Rucaparib is not toxic but significantly enhances temozolomide-induced TGD in the DNA repair protein-competent D384Med xenografts. Pharmacokinetics studies also show that Rucaparib is detected in the brain tissue, which indicates that Rucaparib has potential in intra-cranial malignancy therapy. Rucaparib significantly potentiates the cytotoxicity of topotecan and temozolomide in NB-1691, SH-SY-5Y, and SKNBE (2c) cells. Rucaparib enhances the antitumor activity of temozolomide and indicates complete and sustained tumor regression in NB1691 and SHSY5Y xenografts.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | PAR BRCA1 |
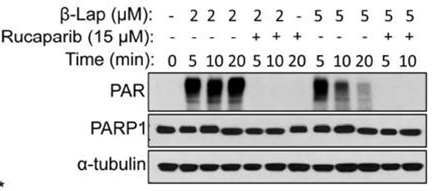
|
27960087 |
| Growth inhibition assay | Cell viability |
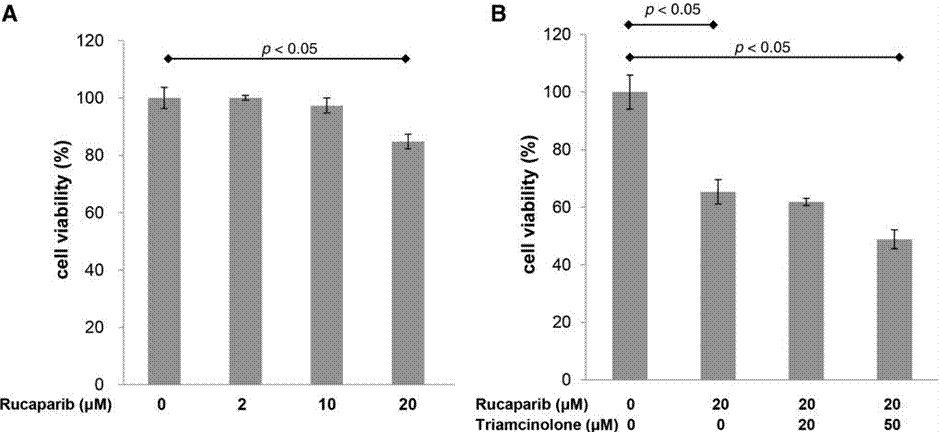
|
31119062 |
| Immunofluorescence | α-tubulin PAR γH2AX 53BP1 |
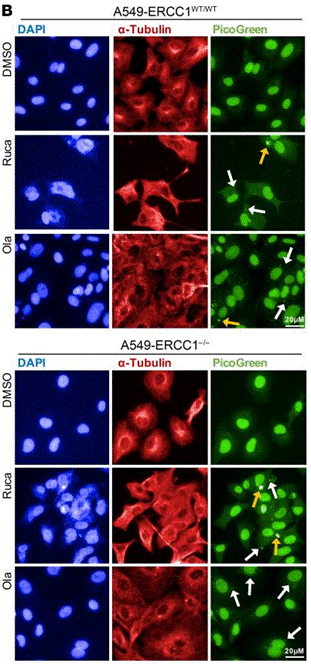
|
30589644 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT04539327 | Completed | Epithelial Ovarian Cancer|Fallopian Tube Cancer|Primary Peritoneal Cancer |
Grupo Español de Investigación en Cáncer de Ovario|Clovis Oncology Inc. |
July 29 2020 | -- |
| NCT04209595 | Active not recruiting | Small Cell Lung Cancer|Extra-Pulmonary Small Cell Carcinomas |
National Cancer Institute (NCI)|National Institutes of Health Clinical Center (CC) |
April 8 2020 | Phase 1|Phase 2 |
| NCT04179396 | Completed | Metastatic Castration Resistant Prostate Cancer |
pharmaand GmbH |
December 5 2019 | Phase 1 |
| NCT03824704 | Terminated | Epithelial Ovarian Cancer|Fallopian Tube Cancer|Primary Peritoneal Carcinoma|High Grade Serous Carcinoma|Endometrioid Adenocarcinoma |
pharmaand GmbH|Bristol-Myers Squibb|Foundation Medicine |
August 23 2019 | Phase 2 |
| NCT03840200 | Completed | Breast Cancer|Prostate Cancer|Ovarian Cancer |
Hoffmann-La Roche |
June 12 2019 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































