research use only
KU-0063794 mTOR inhibitor
Cat.No.S1226
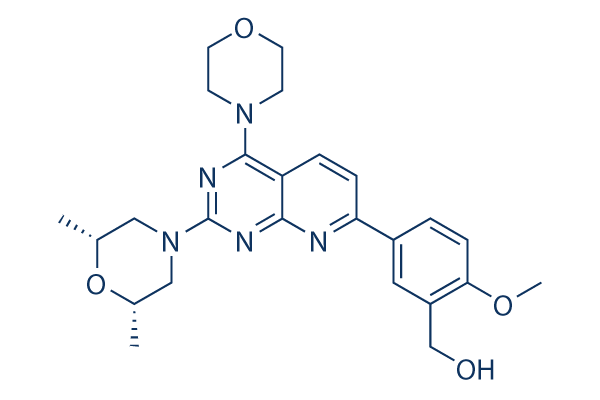
Chemical Structure
Molecular Weight: 465.54
Quality Control
| Related Targets | PI3K Akt GSK-3 ATM/ATR DNA-PK AMPK PDPK1 PTEN PP2A PDK |
|---|---|
| Other mTOR Inhibitors | Torin 1 Torin 2 AZD8055 Ridaforolimus (Deforolimus, MK-8669) Sapanisertib (MLN0128, INK-128) Torkinib (PP242) MHY1485 Vistusertib (AZD2014) OSI-027 3BDO |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| HepG2 | Cell Viability Assay | 0.1–50 μM | 72 h | decreases cell viability in a dose dependent manner | 26278819 | |
| HepG2 | Colony Formation Assay | 1–50 μM | 10 d | decreases the number of viable HepG2 colonies significantly | 26278819 | |
| HepG2 | Apoptosis Assay | 0.1–50 μM | 48 h | induces apoptosis in a dose dependent manner | 26278819 | |
| HepG2 | Function Assay | 5/10 μM | 24 h | dramatically inhibits phosphorylation of AKT at Ser-473 | 26278819 | |
| HepG2 | Function Assay | 5/10 μM | 24 h | downregulates the levels HIF1α and cyclin D1 | 26278819 | |
| HepG2 | Function Assay | 0.1–50 μM | 24 h | induces p62 downregulation, Beclin-1 expression and LC3B-I to LC3B-II conversion in a dose dependent manner | 26278819 | |
| HepG3 | Function Assay | 0.1–50 μM | 48 h | induces cell autophagy in a dose dependent manner | 26278819 | |
| AGS | Growth Inhibition Assay | IC50=15.0 ± 2.91 μM | 24597478 | |||
| HGC27 | Growth Inhibition Assay | IC50=15.0 ± 4.82 μM | 24597478 | |||
| MKN45 | Growth Inhibition Assay | IC50=0.82 ± 0.01 μM | 24597478 | |||
| NUGC4 | Growth Inhibition Assay | IC50=2.93 ± 0.31 μM | 24597478 | |||
| PC9 | Growth Inhibition Assay | 72 h | IC50=10.15±0.62 nM | 23874880 | ||
| PC9GR | Growth Inhibition Assay | 72 h | IC50=6.21±1.30 nM | 23874880 | ||
| H1650 | Growth Inhibition Assay | 72 h | IC50=7.61±0.62 nM | 23874880 | ||
| H1975 | Growth Inhibition Assay | 72 h | IC50=11.15±0.93 nM | 23874880 | ||
| PC9 | Function Assay | 10.15 nM | 72 h | inhibits mTOR phosphorylation status | 23874880 | |
| PC9GR | Function Assay | 6.21 nM | 72 h | inhibits mTOR phosphorylation status | 23874880 | |
| H1650 | Function Assay | 7.61 nM | 72 h | inhibits mTOR phosphorylation status | 23874880 | |
| H1975 | Function Assay | 11.15 nM | 72 h | inhibits mTOR phosphorylation status | 23874880 | |
| PC9 | Function Assay | 10.15 nM | 72 h | inhibits phosphorylation of p70S6K | 23874880 | |
| PC9GR | Function Assay | 6.21 nM | 72 h | inhibits phosphorylation of p70S6K | 23874880 | |
| H1650 | Function Assay | 7.61 nM | 72 h | inhibits phosphorylation of p70S6K | 23874880 | |
| H1975 | Function Assay | 11.15 nM | 72 h | inhibits phosphorylation of p70S6K | 23874880 | |
| LNCaP | Cell Viability Assay | 0-10 μM | 24 h | decreases cell viability in a dose dependent manner | 23840605 | |
| PC-3 | Cell Viability Assay | 0-10 μM | 24 h | decreases cell viability in a dose dependent manner | 23840605 | |
| MDA-MB-468 | Cell Viability Assay | 0-10 μM | 24 h | decreases cell viability in a dose dependent manner | 23840605 | |
| LNCaP | Function Assay | 200–800 nM | 24 h | decreases the phosphorylation level of p70S6K in a dose dependent manner | 23840605 | |
| PC-3 | Function Assay | 200–800 nM | 24 h | decreases the phosphorylation level of p70S6K in a dose dependent manner | 23840605 | |
| MDA-MB-468 | Function Assay | 200–800 nM | 24 h | decreases the phosphorylation level of p70S6K in a dose dependent manner | 23840605 | |
| Caki-1 | Function Assay | 100-2000 nM | 10-180 min | DMSO | inhibits both mTORC1 and mTORC2 as indicated by the decrease in phosphorylation of downstream effectors | 23349989 |
| 786-O | Function Assay | 100-2000 nM | 10-180 min | DMSO | inhibits both mTORC1 and mTORC2 as indicated by the decrease in phosphorylation of downstream effectors | 23349989 |
| Caki-1 | Cell Viability Assay | 300-4000 nM | 24-96 h | DMSO | suppresses the cell viability in both time and dose dependent manner | 23349989 |
| 786-O | Cell Viability Assay | 300-4000 nM | 24-96 h | DMSO | suppresses the cell viability in both time and dose dependent manner | 23349989 |
| Caki-1 | Function Assay | 2 µM | 72 h | DMSO | induces G1 cell cycle arrest and autophagy | 23349989 |
| 786-O | Function Assay | 2 µM | 72 h | DMSO | induces G1 cell cycle arrest and autophagy | 23349989 |
| HEK293 | Function assay | 2 hrs | Inhibition of recombinant FLAG-tagged mTOR expressed in HEK293 cells using biotinylated p70S6K substrate after 2 hrs by alphascreen competition assay, IC50=0.003μM | 19762236 | ||
| HBCx-10 | Function assay | 5 mg/kg | Potentiation of irinotecan-induced tumor regression against human HBCx-10 cells xenografted in immunocompromised mouse at 5 mg/kg, po qd administered on days 1 to 3 of weekly cycle | 19762236 | ||
| U87MG | Function assay | 2 hrs | Inhibition of mTORC1 in human U87MG cells assessed as phosphorylated S6 ribosomal protein (Ser235/236) level after 2 hrs by Western blotting, IC50=0.1μM | 19762236 | ||
| U87MG | Function assay | 2 hrs | Inhibition of mTORC2 in human U87MG cells assessed as phosphorylated AKT (Ser473) level after 2 hrs by Western blotting, IC50=0.15μM | 19762236 | ||
| T47D | Antiproliferative assay | 120 hrs | Antiproliferative activity against human T47D cells after 120 hrs by SRB assay, GI50=0.35μM | 19762236 | ||
| HEK293 | Function assay | Inhibition of recombinant FLAG-tagged mTOR (1362 to 2549) (unknown origin) expressed in HEK293 cells, IC50=0.0025μM | 23375793 | |||
| MDA-MB-468 | Function assay | 2 hrs | Inhibition of mTORC2 in human MDA-MB-468 cells assessed as reduction of AKT phosphorylation at Ser473 after 2 hrs, IC50=0.24μM | 23375793 | ||
| MDA-MB-468 | Function assay | 2 hrs | Inhibition of mTORC1 in human MDA-MB-468 cells assessed as reduction of pS6 phosphorylation at Ser235/236 after 2 hrs, IC50=0.66μM | 23375793 | ||
| HEK293 | Function assay | 30 mins | Inhibition of mTORC1 in HEK293 cells using GST-tagged S6K1 or Akt1 as substrate after 30 mins by immunoblotting assay, IC50=0.01μM | 29211480 | ||
| PC3 | Function assay | 100 mg/kg | Inhibition of Akt phosphorylation at Ser473 in PTEN-deficient human PC3 cells xenograft mouse model at 100 mg/kg, po single dose measured up to 8 hrs | 29211480 | ||
| PC3 | Antitumor assay | 30 mg/kg | Antitumor activity against human PC3 cells xenograft mouse model assessed as inhibition of tumor growth at 30 mg/kg, po bid in presence of 1-aminobenzotriazole | 29211480 | ||
| PC3 | Antitumor assay | 60 mg/kg | Antitumor activity against human PC3 cells xenograft mouse model assessed as inhibition of tumor growth at 60 mg/kg, po bid in presence of 1-aminobenzotriazole | 29211480 | ||
| HEK293 | Function assay | 30 mins | Inhibition of mTORC2 in HEK293 cells using GST-tagged S6K1 or Akt1 as substrate after 30 mins by immunoblotting assay, IC50=0.01μM | 29211480 | ||
| SW620 | Function assay | 20 mg/kg | Potentiation of irinotecan-induced tumor regression against human SW620 cells xenografted in immunocompromised mouse at 20 mg/kg, po qd administered on days 2 to 4 of weekly cycle | 29211480 | ||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh41 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for A673 cells) | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for BT-12 cells | 29435139 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for DAOY cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SJ-GBM2 cells | 29435139 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for U-2 OS cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh41 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for RD cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh18 cells | 29435139 | |||
| Rh30 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh30 cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-SH cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 16 mg/mL
(34.36 mM)
Ethanol : 5 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 465.54 | Formula | C25H31N5O4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 938440-64-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC1CN(CC(O1)C)C2=NC3=C(C=CC(=N3)C4=CC(=C(C=C4)OC)CO)C(=N2)N5CCOCC5 | ||
Mechanism of Action
| Targets/IC50/Ki |
mTORC1
(Cell-free assay) ~10 nM
mTORC2
(Cell-free assay) ~10 nM
|
|---|---|
| In vitro |
Compared with the mTOR inhibitor PP242, KU-0063794 exhibits higher specificity for mTOR, as being inactive against PI3Ks or 76 other kinases. In HEK-293 cells, this compound at 30 nM is sufficient to rapidly ablate S6K1 activity by blocking the phosphorylation of the hydrophobic motif (Thr389) and subsequently the phosphorylation of the T-loop residue (Thr229). In case of IGF1 stimulation of serum-starved HEK-293 cells, 300 nM of this compound is needed to inhibit the S6K1 activity by ~90%. This chemical at 100-300 nM also completely inhibits the amino-acid-induced phosphorylation of S6K1 and S6 protein. Similar to S6K1, it inhibits the phosphorylation of mTORC1 at Ser2448 and mTORC2 at Ser2481 in a dose-dependent and time-dependent manner. In the presence of serum or following IGF1 stimulation, this compound induces a dose-dependent inhibition of the activity and phosphorylation of Akt at Ser473 and unexpected Thr308 as well as the phosphorylation of the Akt substrates PRAS40 at Thr246, GSK3α/GSK3β at Ser21/Ser9 and Foxo-1/3a at Thr24/Thr32. This compound but not rapamycin inhibits SGK1 activity and Ser422 phosphorylation as well as its physiological substrate NDGR1 in a dose-dependent manner, to the same extent as S6K1 and Akt phosphorylation, whereas it dose not inhibit phorbol ester induced ERK or RSK phosphorylation and RSK activation. Compared with rapamycin, this chemical exhibits more significant potency to induce the complete dephosphorylation of 4E-BP1 at Thr37, Thr46 and Ser65. It inhibits cell growth of both wild-type and mLST8-deficient MEFs and induces a G1 cell cycle arrest, more significantly than rapamycin. |
| Kinase Assay |
mTOR complexes kinase assays
|
|
HEK-293 cells are freshly lysed in Hepes lysis buffer. Lysate (1-4 mg) is pre-cleared by incubating with 5-20 μL of Protein G-Sepharose conjugated to pre-immune IgG. The lysate extracts are then incubated with 5-20 μL of Protein G-Sepharose conjugated to 5-20 μg of either anti-Rictor or anti-Raptor antibody, or pre-immune IgG. All antibodies are covalently conjugated to Protein G-Sepharose. Immunoprecipitations are carried out for 1 hour at 4 °C on a vibrating platform. The immunoprecipitates are washed four times with Hepes lysis buffer, followed by two washes with Hepes kinase buffer. For Raptor immunoprecipitates used for phosphorylating S6K1, for the initial two wash steps the buffer includes 0.5 M NaCl to ensure optimal kinase activity. GST-Akt1 is isolated from serum-deprived HEK-293 cells incubated with PI-103 (1 μM for 1 hour). GST-S6K1 is purified from serum-deprived HEK-293 cells incubated with rapamycin (0.1 μM for 1 hour). mTOR reactions are initiated by adding 0.1 mM ATP and 10 mM MgCl2 in the presence of various concentrations of KU-0063794 and GST-Akt1 (0.5 μg) or GST-S6K1 (0.5 μg). Reaction are carried out for 30 minutes at 30 °C on a vibrating platform and stopped by addition of SDS sample buffer. Reaction mixtures are then filtered through a 0.22-μm-poresize Spin-X filter and samples are subjected to electrophoresis and immunoblot analysis with the indicated antibodies.
|
|
| In vivo |
Ku0063794 inhibits tumor growth and mTOR signaling in a preclinical renal cell carcinoma model. However, this compound was not more effective in the animal study. A possible explanation for lack of greater activity in vivo for this chemical. |
References |
|
Applications
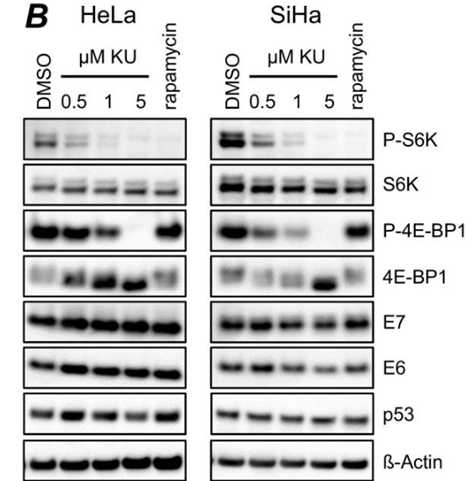
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-S6K / S6K / p-4E-BP1 / E7 / E6 / p53 p-mTOR |
|
28115701 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































