- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- DNA Damage/DNA Repair
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
research use only
VE-821 ATM/ATR inhibitor
VE-821(ATR inhibitor IV) is a potent and selective ATP competitive inhibitor of ATR with Ki/IC50 of 13 nM/26 nM in cell-free assays, shows inhibition of H2AX phosphorylation, minimal activity against PIKKs ATM, DNA-PK, mTOR and PI3Kγ.
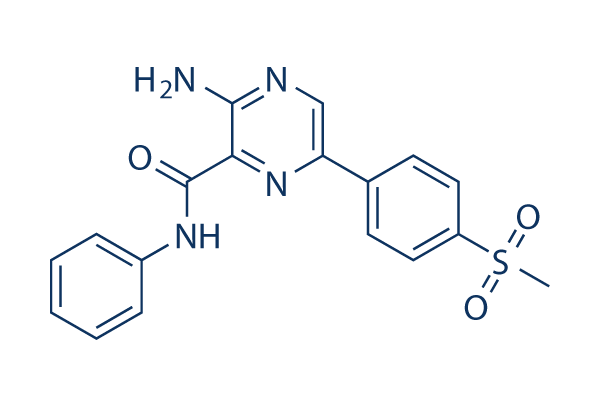
Chemical Structure
Molecular Weight: 368.41
Purity & Quality Control
Batch:
Purity:
99.74%
99.74
Related Products
| Related Targets | ATM ATR | Click to Expand |
|---|---|---|
| Related Products | KU-55933 KU-60019 Berzosertib (VE-822) Ceralasertib (AZD6738) AZ20 AZD0156 Mirin AZD1390 CP-466722 Elimusertib (BAY-1895344) hydrochloride ETP-46464 Elimusertib (BAY-1895344) CGK 733 VX-803 (M4344) AZ31 AZ32 HAMNO | Click to Expand |
| Related Compound Libraries | FDA-approved Drug Library Natural Product Library Apoptosis Compound Library DNA Damage/DNA Repair compound Library Cell Cycle compound library | Click to Expand |
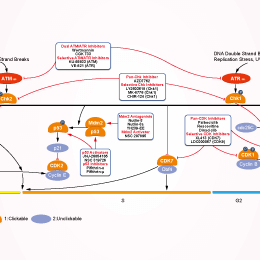
Signaling Pathway
Cell Culture and Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| U2OS | Growth Inhibition Assay | IC50~0.8 μM | 25593184 | |||
| SAOS2 | Growth Inhibition Assay | IC50~0.8 μM | 25593184 | |||
| CAL72 | Growth Inhibition Assay | IC50~0.8 μM | 25593184 | |||
| NOS1 | Growth Inhibition Assay | IC50~0.8 μM | 25593184 | |||
| HUO9 | Growth Inhibition Assay | IC50~0.8 μM | 25593184 | |||
| MG63 | Growth Inhibition Assay | IC50~9 μM | 25593184 | |||
| SJSA1 | Growth Inhibition Assay | IC50~9 μM | 25593184 | |||
| MDA-MB-231 | Cytotoxicity Assay | 1/3/10 μM | 1 h | potentiates the cytotoxicity of both camptothecin and LMP-400 | 25269479 | |
| HT-29 | Cytotoxicity Assay | 1/3/10 μM | 1 h | potentiates the cytotoxicity of both camptothecin and LMP-400 | 25269479 | |
| HCT-116 p53+/+ | Cytotoxicity Assay | 1/3/10 μM | 1 h | potentiates the cytotoxicity of both camptothecin and LMP-400 | 25269479 | |
| HCT-116 p53-/- | Cytotoxicity Assay | 1/3/10 μM | 1 h | potentiates the cytotoxicity of both camptothecin and LMP-400 | 25269479 | |
| TF-1 | Growth Inhibition Assay | 0.011–8 μM | 96 h | enhances the antiproliferative effects of MK1775 | 24179152 | |
| HEL | Growth Inhibition Assay | 0.011–8 μM | 96 h | enhances the antiproliferative effects of MK1775 | 24179152 | |
| THP-1 | Growth Inhibition Assay | 0.011–8 μM | 96 h | enhances the antiproliferative effects of MK1775 | 24179152 | |
| HL-60 | Function Assay | 10 mM | 0.5 h | reduces phosphorylation of Chk1 at serine 345 | 23934411 | |
| OVCAR-8 | Function Assay | 1 µM | 24 h | abrogates chemotherapy-induced cell cycle arrest | 23548269 | |
| PANC-1 | Growth Inhibition Assay | 0.11-9 μM | 1 h | inhibits the cell viability in a dose- dependent manner | 22825331 | |
| MiaPaCa | Growth Inhibition Assay | 0.11-9 μM | 1 h | inhibits the cell viability in a dose- dependent manner | 22825331 | |
| PSN-1 | Growth Inhibition Assay | 0.11-9 μM | 1 h | inhibits the cell viability in a dose- dependent manner | 22825331 | |
| K562 | Apoptosis assay | 10 uM | 48 hrs | Induction of apoptosis in human K562 cells assessed as gammaH2AX/53BP1 levels at 10 uM after 48 hrs by DAPI staining based immunofluorescence microscopy in presence of 25 to 250 pM (-)-lomaiviticin A | 27177826 | |
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| MDCK | Cytotoxicity assay | 500 nM | 1 hr | Cytotoxicity against MDCK cells expressing carbonic anhydrase 9 assessed as reduction in cell viability at 500 nM pretreated for 1 hr followed by 2 Gy irradiation under normoxic condition by Alamar blue assay | 27823879 | |
| MDCK | Cytotoxicity assay | 500 nM | 1 hr | Cytotoxicity against MDCK cells expressing carbonic anhydrase 9 assessed as reduction in cell viability at 500 nM pretreated for 1 hr followed by 4 Gy irradiation under anoxic condition by Alamar blue assay | 27823879 | |
| MDCK | Cytotoxicity assay | 500 nM | 72 hrs | Cytotoxicity against MDCK cells expressing carbonic anhydrase 9 assessed as reduction in cell viability at 500 nM pretreated for 72 hrs followed by 72 hrs incubation under normoxic condition by Alamar blue assay | 27823879 | |
| MDCK | Cytotoxicity assay | 500 nM | 72 hrs | Cytotoxicity against MDCK cells expressing carbonic anhydrase 9 assessed as reduction in cell viability at 500 nM after 72 hrs under anoxic condition by Alamar blue assay | 27823879 | |
| Click to View More Cell Line Experimental Data | ||||||
Mechanism of Action
| Targets |
|
|---|
In vitro |
||||
| In vitro | VE-821(ATR inhibitor IV) shows excellent selectivity for ATR with minimal cross-reactivity against the related PIKKs ATM, DNA-PK, mTOR and PI3K (Kis of 16 μM, 2.2 μM, >1 μM and 3.9 μM, respectively. This compound alone commits a large fraction of cancer cell populations to death, but it only reversibly limits cell cycle progression in normal cells, with minimal death or long-term detrimental effects. [1] This chemical inhibits H2AX cell growth with IC50 of 800 nM. [2] |
|||
|---|---|---|---|---|
| Kinase Assay | Kinase inhibition | |||
| The ability of compounds to inhibit ATR, ATM or DNAPK kinase activity is tested using a radiometric-phosphate incorporation assay. A stock solution is prepared consisting of the appropriate buffer, kinase, and target peptide. To this is added the compound of interest, at varying concentrations in DMSO to a final DMSO concentration of 7%. Assays are initiated by addition of an appropriate [γ-33P]ATP solution and incubated at 25 ℃. Assays are stopped, after the desired time course, by addition of phosphoric acid and ATP to a final concentration of 100 mM and 0.66μM, respectively. Peptides are captured on a phosphocellulose membrane, prepared as per manufacturer | ||||
| Cell Research | Cell lines | H2AX cells | ||
| Concentrations | -- | |||
| Incubation Time | 96 hours | |||
| Method | Cells are plated in 96-well plates and allowed to adhere overnight. The following day, this compound is added at the indicated concentrations in a final volume of 200μL, and the cells are then incubated for 96 h. MTS reagent (40μL) is then added, and 1 h later, absorbance at 490 nm is measured using a SpectraMax Plus 384 plate reader. Synergy and antagonism are assessed using Macsynergy software. |
|||
| Experimental Result Images | Methods | Biomarkers | Images | PMID |
| Western blot | E-cadherin / Vimentin / ZEB1 Vimentin p-AKT / AKT / p-ERK / ERK |
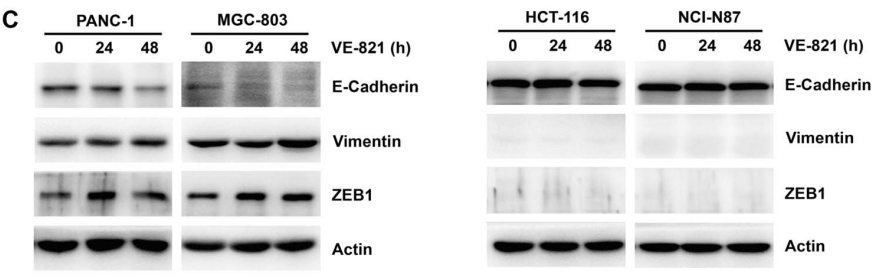
|
29157079 | |
| Growth inhibition assay | Cell viability |
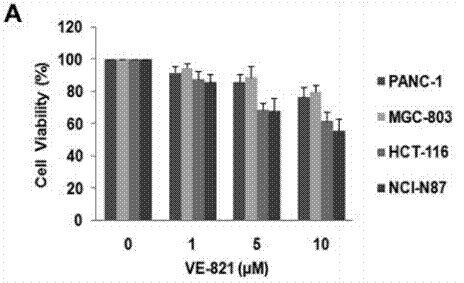
|
29157079 | |
In Vivo |
||
| In vivo | VE-821(ATR inhibitor IV) is a potent and selective ATP competitive inhibitor of ATR. |
|
|---|---|---|
| Animal Research | Animal Models | Male immunodefcient mice |
| Dosages | 15 mg/kg | |
| Administration | i.p. | |
References |
|
Chemical Information
| Molecular Weight | 368.41 | Formula | C18H16N4O3S |
| CAS No. | 1232410-49-9 | SDF | Download SDF |
| Synonyms | ATR inhibitor IV | ||
| Smiles | CS(=O)(=O)C1=CC=C(C=C1)C2=CN=C(C(=N2)C(=O)NC3=CC=CC=C3)N | ||
Storage and Stability
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 74 mg/mL ( (200.86 mM) Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Water : Insoluble Ethanol : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
|||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field