research use only
Piperlongumine ROS chemical
Cat.No.S7551
Chemical Structure
Molecular Weight: 317.34
Quality Control
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| ZR75-30 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human ZR75-30 cells after 72 hrs by MTT assay, IC50=5.86μM | 24937186 | ||
| HCT116 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT116 cells after 72 hrs by MTT assay, IC50=6.04μM | 24937186 | ||
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-231 cells after 72 hrs by MTT assay, IC50=8.46μM | 24937186 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells after 72 hrs by MTT assay, IC50=22.85μM | 24937186 | ||
| WI38 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human WI38 cells after 72 hrs by MTT assay, IC50=26.78μM | 24937186 | ||
| MRC5 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MRC5 cells after 72 hrs by MTT assay, IC50=35.04μM | 24937186 | ||
| PANC1 | Cytotoxicity assay | 24 hrs | Cytotoxicity against human PANC1 cells assessed as reduction in cell viability after 24 hrs by MTT reduction assay, IC50=3.2μM | 25305718 | ||
| HaCaT | Cytotoxicity assay | 24 hrs | Cytotoxicity against human HaCaT cells assessed as reduction in cell viability after 24 hrs by MTT reduction assay, IC50=16μM | 25305718 | ||
| RAW264.7 | Antiinflammatory assay | Antiinflammatory activity in mouse RAW264.7 cells assessed as inhibition of LPS-induced NO production, IC50=3μM | 25453809 | |||
| MA9.3 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MA9.3 cells after 48 hrs by MTT assay, IC50=2μM | 25464887 | ||
| SK-MEL-2 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human SK-MEL-2 cells after 72 hrs by MTT assay, EC50=4.39μM | 25826398 | ||
| HaCaT | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HaCaT cells after 72 hrs by MTT assay, EC50=4.46μM | 25826398 | ||
| A375 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A375 cells after 72 hrs by MTT assay, EC50=6.17μM | 25826398 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by MTT assay, EC50=11.5μM | 25826398 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by MTT assay, EC50=12.2μM | 25826398 | ||
| HFF1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against HFF1 cells after 72 hrs by MTT assay, EC50=13.1μM | 25826398 | ||
| MCF10A | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MCF10A cells after 72 hrs by MTT assay, EC50=18.1μM | 25826398 | ||
| A549 | Function assay | 3 hrs | Induction of ROS production in human A549 cells after 3 hrs by DHE staining-based fluorescence assay | 25826398 | ||
| A549 | Function assay | 3 hrs | Induction of ROS production in human A549 cells after 3 hrs by APF staining-based fluorescence assay | 25826398 | ||
| A549 | Function assay | 10 uM | 4 hrs | Reduction in total GSH level in human A549 cells at 10 uM after 4 hrs by luminescence assay | 25826398 | |
| A549 | Function assay | 10 uM | 4 hrs | Reduction in reduced GSH level in human A549 cells at 10 uM after 4 hrs by luminescence assay | 25826398 | |
| HT1080 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HT1080 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=3.43μM | 26599530 | ||
| GBM10 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human GBM10 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=3.81μM | 26599530 | ||
| SF188 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SF188 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=3.9μM | 26599530 | ||
| SKBR3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SKBR3 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=4.04μM | 26599530 | ||
| T98G | Cytotoxicity assay | 72 hrs | Cytotoxicity against human T98G cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=4.92μM | 26599530 | ||
| HT-29 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HT-29 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=5.49μM | 26599530 | ||
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MCF7 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=6.14μM | 26599530 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells assessed as cell viability after 72 hrs by resazurin reduction assay, IC50=6.54μM | 26599530 | ||
| MV4-11 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MV4-11 cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.2891μM | 27505848 | ||
| CMK | Antiproliferative assay | 72 hrs | Antiproliferative activity against Ara-C resistant human CMK cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.4218μM | 27505848 | ||
| CMY | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMY cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.4424μM | 27505848 | ||
| HL60 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HL60 cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.5292μM | 27505848 | ||
| CMK | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMK cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.6127μM | 27505848 | ||
| U937 | Antiproliferative assay | 72 hrs | Antiproliferative activity against p53 null human U937 cells after 72 hrs in presence of SAHA by MTT assay, IC50=0.767μM | 27505848 | ||
| MOLM13 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MOLM13 cells harboring wild type p53/FLT3-ITD mutant after 72 hrs in presence of SAHA by MTT assay, IC50=0.9345μM | 27505848 | ||
| CMS | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMS cells after 72 hrs in presence of SAHA by MTT assay, IC50=1.037μM | 27505848 | ||
| OCI-AML3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human OCI-AML3 cells after 72 hrs in presence of SAHA by MTT assay, IC50=1.109μM | 27505848 | ||
| HL60 | Antiproliferative assay | 72 hrs | Antiproliferative activity against Ara-C resistant human HL60 cells after 72 hrs in presence of SAHA by MTT assay, IC50=1.128μM | 27505848 | ||
| MV4-11 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MV4-11 cells after 72 hrs by MTT assay, IC50=1.439μM | 27505848 | ||
| CMY | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMY cells after 72 hrs by MTT assay, IC50=1.62μM | 27505848 | ||
| HL60 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HL60 cells after 72 hrs by MTT assay, IC50=1.778μM | 27505848 | ||
| MOLM13 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MOLM13 cells harboring wild type p53/FLT3-ITD mutant after 72 hrs by MTT assay, IC50=1.838μM | 27505848 | ||
| OCI-AML3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human OCI-AML3 cells after 72 hrs by MTT assay, IC50=1.868μM | 27505848 | ||
| CMK | Antiproliferative assay | 72 hrs | Antiproliferative activity against Ara-C resistant human CMK cells after 72 hrs by MTT assay, IC50=1.911μM | 27505848 | ||
| U937 | Antiproliferative assay | 72 hrs | Antiproliferative activity against p53 null human U937 cells after 72 hrs by MTT assay, IC50=2.174μM | 27505848 | ||
| HL60 | Antiproliferative assay | 72 hrs | Antiproliferative activity against Ara-C resistant human HL60 cells after 72 hrs by MTT assay, IC50=2.25μM | 27505848 | ||
| CMK | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMK cells after 72 hrs by MTT assay, IC50=2.815μM | 27505848 | ||
| CMS | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CMS cells after 72 hrs by MTT assay, IC50=3.097μM | 27505848 | ||
| U937 | Apoptosis assay | 2 uM | 24 hrs | Induction of apoptosis in p53 null human U937 cells assessed as caspase 3 cleavage at 2 uM after 24 hrs in presence of SAHA by Western blot analysis | 27505848 | |
| U937 | Apoptosis assay | 2 uM | 24 hrs | Induction of apoptosis in p53 null human U937 cells assessed as PARP-1 cleavage at 2 uM after 24 hrs in presence of SAHA by Western blot analysis | 27505848 | |
| U937 | Cell death assay | 1 to 4 uM | 24 hrs | Induction of cell death in p53 null human U937 cells at 1 to 4 uM after 24 hrs by annexin V-FITC/PI staining based flow cytometry | 27505848 | |
| U937 | Cytotoxicity assay | 0.5 to 5 uM | 24 hrs | Cytotoxicity against p53 null human U937 cells assessed as decrease in cell viability at 0.5 to 5 uM after 24 hrs in presence of SAHA by MTT assay | 27505848 | |
| U937 | Function assay | 2 uM | 20 hrs | Induction of DNA damage in p53 null human U937 cells at 2 uM after 20 hrs in presence of SAHA by SYBR gold staining based alkaline comet assay | 27505848 | |
| MCF7 | Antiproliferative assay | 48 hrs | Antiproliferative activity in human MCF7 cells assessed as cell viability after 48 hrs by alamarBlue assay, IC50=1.2μM | 27689728 | ||
| Jurkat | Antiproliferative assay | 48 hrs | Antiproliferative activity in human Jurkat cells assessed as cell viability after 48 hrs by alamarBlue assay, IC50=1.4μM | 27689728 | ||
| Jurkat | Cytotoxicity assay | 24 hrs | Cytotoxicity against human Jurkat cells after 24 hrs by MTT assay, IC50=5μM | 27689728 | ||
| MCF7 | Cytotoxicity assay | 24 hrs | Cytotoxicity against human MCF7 cells after 24 hrs by MTT assay, IC50=8μM | 27689728 | ||
| MCF7 | Apoptosis assay | 10 uM | 48 hrs | Induction of apoptosis in human MCF7 cells assessed as downregulation of Bcl2 expression at 10 uM after 48 hrs by Western blot method | 27689728 | |
| MCF7 | Apoptosis assay | 10 uM | 72 hrs | Induction of apoptosis in human MCF7 cells assessed as downregulation of Mcl-1 expression at 10 uM after 72 hrs by Western blot method | 27689728 | |
| MCF7 | Function assay | 20 uM | 16 hrs | Disruption of microtubule network in human MCF7 cells at 20 uM after 16 hrs by immunofluorescence analysis | 27689728 | |
| MCF7 | Function assay | 10 uM | 16 hrs | Induction of microtubule depolymerization in human MCF7 cells at 10 uM after 16 hrs by immunofluorescence analysis | 27689728 | |
| MCF7 | Function assay | 1 to 20 uM | 48 hrs | Induction of reactive oxygen species generation in human MCF7 cells assessed as decrease in cell viability at 1 to 20 uM in presence of trolox after 48 hrs by alamarBlue assay | 27689728 | |
| MCF7 | Function assay | 5 to 20 uM | 4 hrs | Induction of tubulin depolymerization in human MCF7 cells at 5 to 20 uM after 4 hrs by Western blot method | 27689728 | |
| Jurkat | Function assay | 5 to 20 uM | 4 hrs | Induction of tubulin depolymerization in human Jurkat cells at 5 to 20 uM after 4 hrs by Western blot method | 27689728 | |
| A549 | Function assay | 24 hrs | Inhibition of NF-kappaB in human A549 cells after 24 hrs by luciferase reporter gene assay, IC50=1.76μM | 27810594 | ||
| HepG2 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HepG2 cells assessed as growth inhibition after 72 hrs by MTT assay, IC50=8.9μM | 28159415 | ||
| HCT116 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT116 cells assessed as growth inhibition after 72 hrs by MTT assay, IC50=9.2μM | 28159415 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells assessed as growth inhibition after 72 hrs by MTT assay, IC50=10μM | 28159415 | ||
| MRC5 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MRC5 cells after 72 hrs by MTT assay, IC50=37μM | 28159415 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by MTT assay, IC50=6.84μM | 28434764 | ||
| Saos2 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human Saos2 cells after 72 hrs by MTT assay, IC50=7.31μM | 28434764 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by MTT assay, IC50=7.34μM | 28434764 | ||
| U2OS | Antiproliferative assay | 72 hrs | Antiproliferative activity against human U2OS cells after 72 hrs by MTT assay, IC50=9.49μM | 28434764 | ||
| MDA-MB-231 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MDA-MB-231 cells after 72 hrs by MTT assay, IC50=10.6μM | 28434764 | ||
| SKHEP1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human SKHEP1 cells after 72 hrs by MTT assay, IC50=13.3μM | 28434764 | ||
| HCT116 | Function assay | 10 uM | 1 hr | Induction of reactive oxygen species generation in human HCT116 cells at 10 uM after 1 hr by DCFH-DA dye-based fluorescence microscopic analysis | 28434764 | |
| HCT116 | Cell cycle assay | 1 to 5 uM | 24 hrs | Cell cycle arrest in human HCT116 cells assessed as accumulation at S phase 1 to 5 uM after 24 hrs by propidium iodide staining based flow cytometric method | 28434764 | |
| HCT116 | Cell cycle assay | 1 to 5 uM | 24 hrs | Cell cycle arrest in human HCT116 cells assessed as accumulation at G0/G1 phase at 1 to 5 uM after 24 hrs by propidium iodide staining based flow cytometric method | 28434764 | |
| HCT116 | Cell cycle assay | 1 to 5 uM | 24 hrs | Cell cycle arrest in human HCT116 cells decrease in population at G0/G1 phase at 1 to 5 uM after 24 hrs by propidium iodide staining based flow cytometric method | 28434764 | |
| HT-29 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HT-29 cells after 72 hrs by MTT assay, IC50=2.65μM | 28686911 | ||
| HCT8 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT8 cells after 72 hrs by MTT assay, IC50=4.1μM | 28686911 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by MTT assay, IC50=8.13μM | 28686911 | ||
| CCD-841 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CCD-841 cells assessed as reduction in cell viability after 72 hrs by MTT assay, IC50=44.32μM | 28686911 | ||
| HCT116 | Antiproliferative assay | 10 uM | 72 hrs | Antiproliferative activity against human HCT116 cells at 10 uM after 72 hrs by MTT assay | 28686911 | |
| A549 | Antiproliferative assay | 10 uM | 72 hrs | Antiproliferative activity against human A549 cells at 10 uM after 72 hrs by MTT assay | 28686911 | |
| K562 | Antiproliferative assay | 10 uM | 72 hrs | Antiproliferative activity against human K562 cells at 10 uM after 72 hrs by MTT assay | 28686911 | |
| HT-29 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HT-29 cells after 72 hrs by MTT assay, IC50=2.65μM | 29424539 | ||
| HCT8 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT8 cells after 72 hrs by MTT assay, IC50=4.1μM | 29424539 | ||
| SW620 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human SW620 cells after 72 hrs by MTT assay, IC50=4.62μM | 29424539 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells after 72 hrs by MTT assay, IC50=5.05μM | 29424539 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells after 72 hrs in presence of ligustrazine by MTT assay, IC50=5.09μM | 29424539 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by MTT assay, IC50=8.13μM | 29424539 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs in presence of ligustrazine by MTT assay, IC50=8.17μM | 29424539 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs in presence of ligustrazine by MTT assay, IC50=15.22μM | 29424539 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by MTT assay, IC50=15.28μM | 29424539 | ||
| U87MG | Antiproliferative assay | 72 hrs | Antiproliferative activity against human U87MG cells after 72 hrs in presence of ligustrazine by MTT assay, IC50=16.15μM | 29424539 | ||
| U87MG | Antiproliferative assay | 72 hrs | Antiproliferative activity against human U87MG cells after 72 hrs by MTT assay, IC50=17.34μM | 29424539 | ||
| CCD-841 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CCD-841 cells after 72 hrs by MTT assay, IC50=44.32μM | 29424539 | ||
| HCT116 | Antimigratory assay | 50 nM | 48 hrs | Antimigratory activity in human HCT116 cells assessed as inhibition of TGF-beta1-induced cell migration at 50 nM after 48 hrs by trans-well assay | 29424539 | |
| HCT116 | Function assay | 10 uM | 3 hrs | Induction of ROS accumulation in human HCT116 cells at 10 uM after 3 hrs by DCFH-DA staining-based fluorescence microscopic analysis | 29424539 | |
| HCT116 | Antiproliferative assay | 8 uM | 72 hrs | Antiproliferative activity against human HCT116 cells at 8 uM after 72 hrs by MTT assay | 29424539 | |
| HCT116 | Antiinvasive assay | 50 nM | 48 hrs | Antiinvasive activity in human HCT116 cells assessed as inhibition of TGF-beta1-induced cell invasion at 50 nM after 48 hrs by crystal violet staining-based assay | 29424539 | |
| HCT116 | Function assay | 50 nM | 1 hr | Inhibition of IL-1beta-mediated adhesion of human rhodamine 123-labeled HCT116 cells to HUVEC at 50 nM after 1 hr by fluoresence assay | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced decrease in expression levels of E-cadherin protein in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced increase in expression levels of vimentin protein in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced increase in expression levels of MMP-9 protein in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced increase in expression levels of snail protein in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced increase in expression levels of twist protein in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced AKT phosphorylation in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-induced GSK-3beta phosphorylation in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-regulated decrease in cytoplasmic beta-catenin in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| HCT116 | Function assay | 50 nM | 48 hrs | Inhibition of TGF-beta1-regulated increase in nuclear beta-catenin in human HCT116 cells at 50 nM after 48 hrs by Western blot analysis | 29424539 | |
| DU145 | Function assay | 24 hrs | Inhibition of STAT3 phosphorylation at Tyr705 residues in human DU145 cells after 24 hrs by Western blot analysis, IC50=7μM | 29807795 | ||
| DU145 | Growth inhibition assay | 48 hrs | Growth inhibition of human DU145 cells after 48 hrs by WST-1 assay, GI50=7.1μM | 29807795 | ||
| LNCAP | Growth inhibition assay | 48 hrs | Growth inhibition of human LNCAP cells after 48 hrs by WST-1 assay, GI50=9.2μM | 29807795 | ||
| WI38 | Function assay | Senolytic activity against ionizing radiation induced human WI38 senescent cells assessed as reduction in cell viability by propidium iodide staining based flow cytometry, EC50=8μM | 29925484 | |||
| WI38 | Function assay | Senolytic activity against non-senescent human WI38 cells assessed as reduction in cell viability by propidium iodide staining based flow cytometry, EC50=20.3μM | 29925484 | |||
| WI38 | Function assay | 5 uM | 24 hrs | Induction of OXR1 degradation in ionizing radiation induced human WI38 senescent cells at 5 uM after 24 hrs by Western blot analysis | 29925484 | |
| WI38 | Function assay | up to 64 uM | 1.5 hrs | Induction of ROS production in ionizing radiation induced human WI38 senescent cells up to 64 uM after 1.5 hrs by DHR123 dye based flow cytometry | 29925484 | |
| RAW264.7 | Function assay | 10 uM | 0.5 hrs | Inhibition of MAPK/NFkappaB in ICR mouse RAW264.7 cells assessed as reduction in LPS-induced TNFalpha production at 10 uM pretreated for 0.5 hrs followed by LPS challenge and measured after 24 hrs by ELISA relative to control | 30780088 | |
| RAW264.7 | Function assay | 10 uM | 0.5 hrs | Inhibition of MAPK/NFkappaB in ICR mouse RAW264.7 cells assessed as reduction in LPS-induced IL6 production at 10 uM pretreated for 0.5 hrs followed by LPS challenge and measured after 24 hrs by ELISA relative to control | 30780088 | |
| BHK21 | Cytotoxicity assay | 48 hrs | Cytotoxicity against BHK21 cells assessed as reduction in cell viability after 48 hrs by MTT assay, CC50=40.14μM | 31009908 | ||
| SH-SY5Y | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SH-SY5Y cells assessed as reduction in cell viability incubated for 72 hrs by MTT assay, IC50=5.94μM | 31400708 | ||
| U87 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human U87 cells assessed as reduction in cell viability incubated for 72 hrs by MTT assay, IC50=7.18μM | 31400708 | ||
| SH-SY5Y | Function assay | 15 mins | Inhibition of TrxR in human SH-SY5Y cells using DTNB as substrate preincubated for 15 mins followed by substrate addition and measured for 20 mins by Ellman's method, IC50=45.88μM | 31400708 | ||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 63 mg/mL
(198.52 mM)
Ethanol : 63 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 317.34 | Formula | C17H19NO5 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 20069-09-4 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | PPLGM, Piplartine | Smiles | COC1=CC(=CC(=C1OC)OC)C=CC(=O)N2CCC=CC2=O | ||
Mechanism of Action
| Targets/IC50/Ki |
reactive oxygen species (ROS)
TrxR1
CRM1
PI3K/Akt/mTOR
|
|---|---|
| In vitro |
Piperlongumine is a known ROS inducer which could induce pancreatic cancer cell death in cell culture As a thromboxane A(2) receptor antagonist, this compound inhibits platelet aggregation. It also promotes autophagy via inhibition of Akt/mTOR signalling and mediates cancer cell death.
|
| In vivo |
Piperlongumine (50 mg/kg i.p.) causes in vivo growth inhibition of tumor cells without leading to major changes in the biochemical, hematological and histopathological parameters.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | CDK2 / Cyclin E / Cyclin A Bax / Bcl-2 Survivin / p21 / p27 p-STAT3 / STAT3 / p-JAK2 / JAK2 / c-myc Hexokinase 1 / Hexokinase 2 p-AKT / AKT / p-S6 / S6 |
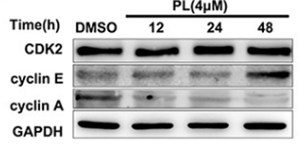
|
27634873 |
| Growth inhibition assay | Cell viability |
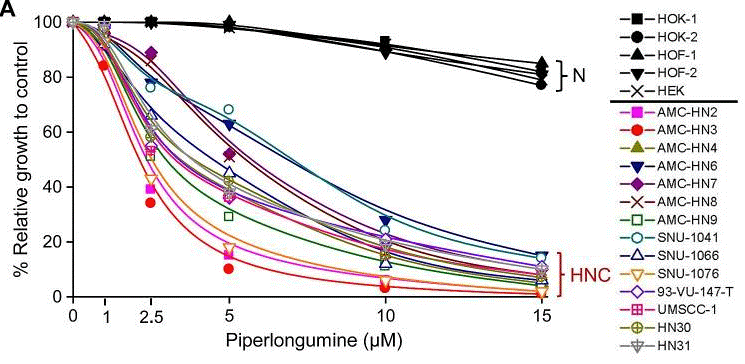
|
25193861 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































