- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
A-769662 AMPK activator
Cat.No.S2697
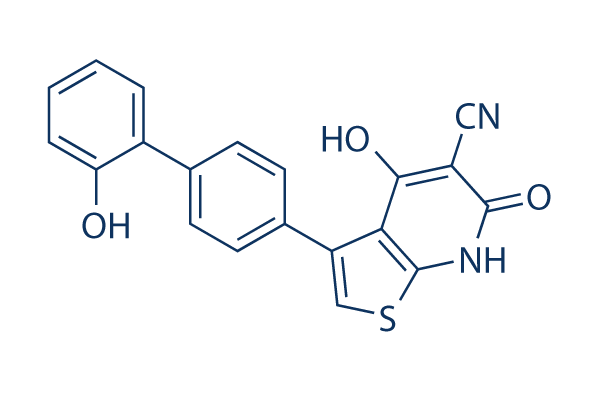
Chemical Structure
Molecular Weight: 360.39
Jump to
Quality Control
Batch:
Purity:
99.67%
99.67
| Related Targets | PI3K Akt mTOR GSK-3 ATM/ATR DNA-PK PDPK1 PTEN PP2A PDK |
|---|---|
| Other AMPK Inhibitors | Dorsomorphin Dihydrochloride Dorsomorphin (Compound C) AICAR (Acadesine) GSK621 WZ4003 ex229 (compound 991) Phenformin HCl HTH-01-015 BAY-3827 O-304 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| mouse hepatocytes | Function assay | 1 mM | DMSO | inhibits fatty acid synthesis with IC50 of 3.6 μM | 16753576 | |
| rat hepatocytes | Function assay | 1 mM | DMSO | inhibits fatty acid synthesis with IC50 of 3.6 μM | 16753576 | |
| HEK293 | Kinase assay | 200 μM | DMSO | activates endogenous AMPK | 17728241 | |
| CCL13 | Kinase assay | 200 μM | DMSO | activates endogenous AMPK | 17728241 | |
| MEFs | Function assay | 300 μM | DMSO | inhibits proteasomal function by an AMPK-independent mechanism | 18593584 | |
| epididymal clear cells | Function assay | 200 μM | DMSO | inhibits the pH-mediated V-ATPase accumulation at the apical membrane | 19211918 | |
| 3T3-L1 | Function assay | 1.2 mM | DMSO | inhibits 3T3-L1 Adipogenesis | 19483304 | |
| 3T3-L1 | Function assay | 1.2 mM | DMSO | inhibits the Expression of AdipogenesisRelated Transcription Factors and Markers | 19483304 | |
| 3T3-L1 | Function assay | 1.2 mM | DMSO | inhibits Mitotic Clonal Expansion | 19483304 | |
| 3T3-L1 | cytotoxicity assay | 1.2 mM | DMSO | decreases Cell Viability | 19483304 | |
| 3T3-L1 | Kinase assay | 1.2 mM | DMSO | activates AMPK | 19483304 | |
| L6 skeletal muscle cells | Function assay | 250 μM | DMSO | activates AMPK signaling pathways | 19828836 | |
| L6 skeletal muscle cells | Function assay | 250 μM | DMSO | inhibits the Na+-K+-ATPase transport activity and cell surface abundance | 19828836 | |
| MDA-MB231 | Apoptosis assay | 400 μM | DMSO | sensitizes human breast cancer cell lines to TRAIL-induced apoptosis | 19896469 | |
| BT474 | Apoptosis assay | 400 μM | DMSO | sensitizes human breast cancer cell lines to TRAIL-induced apoptosis | 19896469 | |
| MCF7 | Apoptosis assay | 400 μM | DMSO | sensitizes human breast cancer cell lines to TRAIL-induced apoptosis | 19896469 | |
| Mesenchymal stem cells | Kinase assay | 10 µM | DMSO | induces a robust and sustained AMPK activation | 24104879 | |
| Mesenchymal stem cells | cytotoxicity assay | 100 µM | DMSO | decreases the MSC proliferation | 24104879 | |
| MG-63 | cytotoxicity assay | 10 µM | DMSO | inhibits H2O2-Induced Osteoblast Cell Death | 24960362 | |
| MC3T3-E1 | cytotoxicity assay | 10 µM | DMSO | inhibits H2O2-Induced Osteoblast Cell Death | 24960362 | |
| MG-63 | Apoptosis assay | 10 µM | DMSO | suppresses H2O2-Induced Osteoblast Cell Apoptosis | 24960362 | |
| MC3T3-E1 | Apoptosis assay | 10 µM | DMSO | suppresses H2O2-Induced Osteoblast Cell Apoptosis | 24960362 | |
| MG-63 | Function assay | 10 µM | DMSO | alleviates ROS accumulation and ATP depletion caused by H2O2 | 24960362 | |
| MC3T3-E1 | Function assay | 10 µM | DMSO | alleviates ROS accumulation and ATP depletion caused by H2O2 | 24960362 | |
| MG-63 | Function assay | 10 µM | DMSO | facilitates H2O2-induced autophagy activation | 24960362 | |
| MC3T3-E1 | Function assay | 10 µM | DMSO | facilitates H2O2-induced autophagy activation | 24960362 | |
| PC3 | Kinase assay | 100 µM | DMSO | upregulates the levels of AMPK and ACC phosphorylation | 25594043 | |
| PC3M | Kinase assay | 100 µM | DMSO | upregulates the levels of AMPK and ACC phosphorylation | 25594043 | |
| PC3 | Function assay | 100 µM | DMSO | induces PI3K/mTOR pathways | 25594043 | |
| PC3M | Function assay | 100 µM | DMSO | induces PI3K/mTOR pathways | 25594043 | |
| PC3 | Growth inhibitory assay | 100 µM | DMSO | suppresses proliferation | 25594043 | |
| PC3M | Growth inhibitory assay | 100 µM | DMSO | suppresses proliferation | 25594043 | |
| MC3T3-E1 | Kinase assay | 10 μM | DMSO | induces significant AMPK activation | 26891866 | |
| MC3T3-E1 | Growth inhibitory assay | 10 μM | DMSO | inhibits Dex-induced osteoblast cell death | 26891866 | |
| MC3T3-E1 | Function assay | 10 μM | DMSO | inhibits Dex-induced oxidative stress | 26891866 | |
| BL21 | Function assay | 1 to 10 mins | Binding affinity to human recombinant N-terminal AVI-tagged phosphorylated AMPK alpha1beta1gamma1 expressed in Escherichia coli BL21 cells after 1 to 10 mins by biolayer interferometry, Kd = 0.51 μM. | 26510622 | ||
| BL21 | Function assay | 1 to 10 mins | Binding affinity to human recombinant N-terminal AVI-tagged phosphorylated AMPK alpha1beta2gamma1 expressed in Escherichia coli BL21 cells after 1 to 10 mins by biolayer interferometry, Kd = 14.5 μM. | 26510622 | ||
| HeLa | Function assay | 100 uM | 1 hr | Effect on AMPK activation in HeLa cells at 100 uM after 1 hr by immunoprecipitate kinase assay | 17855357 | |
| HeLa | Function assay | 100 uM | 1 hr | Effect on AMPK phosphorylation in HeLa cells at 100 uM after 1 hr by immunoprecipitate kinase assay | 17855357 | |
| HeLa | Function assay | 100 uM | 5 mins | Effect on AMPK phosphorylation in HeLa cells at 100 uM after 5 mins by immunoprecipitate kinase assay | 17855357 | |
| HeLa | Function assay | 100 uM | 1 hr | Effect on acetyl-CoA carboxylase phosphorylation in HeLa cells at 100 uM after 1 hr by densitometry | 17855357 | |
| HeLa | Function assay | 100 uM | 1 hr | Reduction of AMPK phosphorylation in HeLa cells at 100 uM after 1 hr in presence of STO-609 by immunoprecipitate kinase assay | 17855357 | |
| HeLa | Function assay | 100 uM | 1 hr | Reduction of AMPK activation in HeLa cells at 100 uM after 1 hr in presence of STO-609 by immunoprecipitate kinase assay | 17855357 | |
| HeLa | Function assay | 100 uM | 1 hr | Reduction of acetyl-CoA carboxylase phosphorylation in HeLa cells at 100 uM after 1 hr in presence of STO-609 by densitometry | 17855357 | |
| CMK | Growth Inhibition Assay | Inhibition of human CMK cell growth in a cell viability assay, IC50 = 0.002202 μM. | SANGER | |||
| SIG-M5 | Growth Inhibition Assay | Inhibition of human SIG-M5 cell growth in a cell viability assay, IC50 = 0.007856 μM. | SANGER | |||
| MV-4-11 | Growth Inhibition Assay | Inhibition of human MV-4-11 cell growth in a cell viability assay, IC50 = 0.02274 μM. | SANGER | |||
| GDM-1 | Growth Inhibition Assay | Inhibition of human GDM-1 cell growth in a cell viability assay, IC50 = 0.02553 μM. | SANGER | |||
| A431 | Growth Inhibition Assay | Inhibition of human A431 cell growth in a cell viability assay, IC50 = 0.02919 μM. | SANGER | |||
| NALM-6 | Growth Inhibition Assay | Inhibition of human NALM-6 cell growth in a cell viability assay, IC50 = 0.03166 μM. | SANGER | |||
| KM12 | Growth Inhibition Assay | Inhibition of human KM12 cell growth in a cell viability assay, IC50 = 0.03213 μM. | SANGER | |||
| QIMR-WIL | Growth Inhibition Assay | Inhibition of human QIMR-WIL cell growth in a cell viability assay, IC50 = 0.03642 μM. | SANGER | |||
| NH-12 | Growth Inhibition Assay | Inhibition of human NH-12 cell growth in a cell viability assay, IC50 = 0.03689 μM. | SANGER | |||
| KASUMI-1 | Growth Inhibition Assay | Inhibition of human KASUMI-1 cell growth in a cell viability assay, IC50 = 0.0418 μM. | SANGER | |||
| ML-2 | Growth Inhibition Assay | Inhibition of human ML-2 cell growth in a cell viability assay, IC50 = 0.04344 μM. | SANGER | |||
| YT | Growth Inhibition Assay | Inhibition of human YT cell growth in a cell viability assay, IC50 = 0.0467 μM. | SANGER | |||
| Daudi | Growth Inhibition Assay | Inhibition of human Daudi cell growth in a cell viability assay, IC50 = 0.04979 μM. | SANGER | |||
| OCI-AML2 | Growth Inhibition Assay | Inhibition of human OCI-AML2 cell growth in a cell viability assay, IC50 = 0.0572 μM. | SANGER | |||
| ALL-PO | Growth Inhibition Assay | Inhibition of human ALL-PO cell growth in a cell viability assay, IC50 = 0.05793 μM. | SANGER | |||
| MOLT-16 | Growth Inhibition Assay | Inhibition of human MOLT-16 cell growth in a cell viability assay, IC50 = 0.06191 μM. | SANGER | |||
| EHEB | Growth Inhibition Assay | Inhibition of human EHEB cell growth in a cell viability assay, IC50 = 0.06325 μM. | SANGER | |||
| AM-38 | Growth Inhibition Assay | Inhibition of human AM-38 cell growth in a cell viability assay, IC50 = 0.0644 μM. | SANGER | |||
| C8166 | Growth Inhibition Assay | Inhibition of human C8166 cell growth in a cell viability assay, IC50 = 0.06619 μM. | SANGER | |||
| RPMI-6666 | Growth Inhibition Assay | Inhibition of human RPMI-6666 cell growth in a cell viability assay, IC50 = 0.06798 μM. | SANGER | |||
| BC-1 | Growth Inhibition Assay | Inhibition of human BC-1 cell growth in a cell viability assay, IC50 = 0.06973 μM. | SANGER | |||
| CTV-1 | Growth Inhibition Assay | Inhibition of human CTV-1 cell growth in a cell viability assay, IC50 = 0.07303 μM. | SANGER | |||
| HC-1 | Growth Inhibition Assay | Inhibition of human HC-1 cell growth in a cell viability assay, IC50 = 0.08314 μM. | SANGER | |||
| JiyoyeP-2003 | Growth Inhibition Assay | Inhibition of human JiyoyeP-2003 cell growth in a cell viability assay, IC50 = 0.08371 μM. | SANGER | |||
| RS4-11 | Growth Inhibition Assay | Inhibition of human RS4-11 cell growth in a cell viability assay, IC50 = 0.08572 μM. | SANGER | |||
| PF-382 | Growth Inhibition Assay | Inhibition of human PF-382 cell growth in a cell viability assay, IC50 = 0.0859 μM. | SANGER | |||
| G-361 | Growth Inhibition Assay | Inhibition of human G-361 cell growth in a cell viability assay, IC50 = 0.08815 μM. | SANGER | |||
| LOXIMVI | Growth Inhibition Assay | Inhibition of human LOXIMVI cell growth in a cell viability assay, IC50 = 0.08833 μM. | SANGER | |||
| A3-KAW | Growth Inhibition Assay | Inhibition of human A3-KAW cell growth in a cell viability assay, IC50 = 0.09169 μM. | SANGER | |||
| EoL-1-cell | Growth Inhibition Assay | Inhibition of human EoL-1-cell cell growth in a cell viability assay, IC50 = 0.09558 μM. | SANGER | |||
| LOUCY | Growth Inhibition Assay | Inhibition of human LOUCY cell growth in a cell viability assay, IC50 = 0.09641 μM. | SANGER | |||
| HL-60 | Growth Inhibition Assay | Inhibition of human HL-60 cell growth in a cell viability assay, IC50 = 0.09739 μM. | SANGER | |||
| MC116 | Growth Inhibition Assay | Inhibition of human MC116 cell growth in a cell viability assay, IC50 = 0.10014 μM. | SANGER | |||
| CESS | Growth Inhibition Assay | Inhibition of human CESS cell growth in a cell viability assay, IC50 = 0.1011 μM. | SANGER | |||
| REH | Growth Inhibition Assay | Inhibition of human REH cell growth in a cell viability assay, IC50 = 0.10382 μM. | SANGER | |||
| HCC2157 | Growth Inhibition Assay | Inhibition of human HCC2157 cell growth in a cell viability assay, IC50 = 0.10455 μM. | SANGER | |||
| HEL | Growth Inhibition Assay | Inhibition of human HEL cell growth in a cell viability assay, IC50 = 0.10728 μM. | SANGER | |||
| BL-70 | Growth Inhibition Assay | Inhibition of human BL-70 cell growth in a cell viability assay, IC50 = 0.10805 μM. | SANGER | |||
| NCI-H1770 | Growth Inhibition Assay | Inhibition of human NCI-H1770 cell growth in a cell viability assay, IC50 = 0.10865 μM. | SANGER | |||
| SK-MM-2 | Growth Inhibition Assay | Inhibition of human SK-MM-2 cell growth in a cell viability assay, IC50 = 0.10944 μM. | SANGER | |||
| MONO-MAC-6 | Growth Inhibition Assay | Inhibition of human MONO-MAC-6 cell growth in a cell viability assay, IC50 = 0.11372 μM. | SANGER | |||
| SR | Growth Inhibition Assay | Inhibition of human SR cell growth in a cell viability assay, IC50 = 0.11387 μM. | SANGER | |||
| MOLT-4 | Growth Inhibition Assay | Inhibition of human MOLT-4 cell growth in a cell viability assay, IC50 = 0.11446 μM. | SANGER | |||
| KE-37 | Growth Inhibition Assay | Inhibition of human KE-37 cell growth in a cell viability assay, IC50 = 0.12924 μM. | SANGER | |||
| ST486 | Growth Inhibition Assay | Inhibition of human ST486 cell growth in a cell viability assay, IC50 = 0.13527 μM. | SANGER | |||
| HDLM-2 | Growth Inhibition Assay | Inhibition of human HDLM-2 cell growth in a cell viability assay, IC50 = 0.13616 μM. | SANGER | |||
| ARH-77 | Growth Inhibition Assay | Inhibition of human ARH-77 cell growth in a cell viability assay, IC50 = 0.1406 μM. | SANGER | |||
| EB2 | Growth Inhibition Assay | Inhibition of human EB2 cell growth in a cell viability assay, IC50 = 0.14234 μM. | SANGER | |||
| CCRF-CEM | Growth Inhibition Assay | Inhibition of human CCRF-CEM cell growth in a cell viability assay, IC50 = 0.14576 μM. | SANGER | |||
| HD-MY-Z | Growth Inhibition Assay | Inhibition of human HD-MY-Z cell growth in a cell viability assay, IC50 = 0.14791 μM. | SANGER | |||
| BT-474 | Growth Inhibition Assay | Inhibition of human BT-474 cell growth in a cell viability assay, IC50 = 0.15074 μM. | SANGER | |||
| KARPAS-45 | Growth Inhibition Assay | Inhibition of human KARPAS-45 cell growth in a cell viability assay, IC50 = 0.16984 μM. | SANGER | |||
| Ramos-2G6-4C10 | Growth Inhibition Assay | Inhibition of human Ramos-2G6-4C10 cell growth in a cell viability assay, IC50 = 0.1715 μM. | SANGER | |||
| MC-CAR | Growth Inhibition Assay | Inhibition of human MC-CAR cell growth in a cell viability assay, IC50 = 0.17153 μM. | SANGER | |||
| MRK-nu-1 | Growth Inhibition Assay | Inhibition of human MRK-nu-1 cell growth in a cell viability assay, IC50 = 0.17751 μM. | SANGER | |||
| MHH-PREB-1 | Growth Inhibition Assay | Inhibition of human MHH-PREB-1 cell growth in a cell viability assay, IC50 = 0.18242 μM. | SANGER | |||
| SK-NEP-1 | Growth Inhibition Assay | Inhibition of human SK-NEP-1 cell growth in a cell viability assay, IC50 = 0.18514 μM. | SANGER | |||
| BL-41 | Growth Inhibition Assay | Inhibition of human BL-41 cell growth in a cell viability assay, IC50 = 0.18943 μM. | SANGER | |||
| CAS-1 | Growth Inhibition Assay | Inhibition of human CAS-1 cell growth in a cell viability assay, IC50 = 0.19307 μM. | SANGER | |||
| NCI-H1522 | Growth Inhibition Assay | Inhibition of human NCI-H1522 cell growth in a cell viability assay, IC50 = 0.19355 μM. | SANGER | |||
| CA46 | Growth Inhibition Assay | Inhibition of human CA46 cell growth in a cell viability assay, IC50 = 0.19556 μM. | SANGER | |||
| L-363 | Growth Inhibition Assay | Inhibition of human L-363 cell growth in a cell viability assay, IC50 = 0.21121 μM. | SANGER | |||
| NCI-H2087 | Growth Inhibition Assay | Inhibition of human NCI-H2087 cell growth in a cell viability assay, IC50 = 0.21446 μM. | SANGER | |||
| GR-ST | Growth Inhibition Assay | Inhibition of human GR-ST cell growth in a cell viability assay, IC50 = 0.21764 μM. | SANGER | |||
| OPM-2 | Growth Inhibition Assay | Inhibition of human OPM-2 cell growth in a cell viability assay, IC50 = 0.21791 μM. | SANGER | |||
| U-698-M | Growth Inhibition Assay | Inhibition of human U-698-M cell growth in a cell viability assay, IC50 = 0.21915 μM. | SANGER | |||
| CAL-148 | Growth Inhibition Assay | Inhibition of human CAL-148 cell growth in a cell viability assay, IC50 = 0.22342 μM. | SANGER | |||
| L-428 | Growth Inhibition Assay | Inhibition of human L-428 cell growth in a cell viability assay, IC50 = 0.22499 μM. | SANGER | |||
| DEL | Growth Inhibition Assay | Inhibition of human DEL cell growth in a cell viability assay, IC50 = 0.22825 μM. | SANGER | |||
| LU-134-A | Growth Inhibition Assay | Inhibition of human LU-134-A cell growth in a cell viability assay, IC50 = 0.23116 μM. | SANGER | |||
| MLMA | Growth Inhibition Assay | Inhibition of human MLMA cell growth in a cell viability assay, IC50 = 0.23399 μM. | SANGER | |||
| DOHH-2 | Growth Inhibition Assay | Inhibition of human DOHH-2 cell growth in a cell viability assay, IC50 = 0.24513 μM. | SANGER | |||
| RPMI-8226 | Growth Inhibition Assay | Inhibition of human RPMI-8226 cell growth in a cell viability assay, IC50 = 0.24596 μM. | SANGER | |||
| NCI-H2196 | Growth Inhibition Assay | Inhibition of human NCI-H2196 cell growth in a cell viability assay, IC50 = 0.24776 μM. | SANGER | |||
| COR-L279 | Growth Inhibition Assay | Inhibition of human COR-L279 cell growth in a cell viability assay, IC50 = 0.25293 μM. | SANGER | |||
| CAPAN-1 | Growth Inhibition Assay | Inhibition of human CAPAN-1 cell growth in a cell viability assay, IC50 = 0.25589 μM. | SANGER | |||
| NOMO-1 | Growth Inhibition Assay | Inhibition of human NOMO-1 cell growth in a cell viability assay, IC50 = 0.26322 μM. | SANGER | |||
| SUP-T1 | Growth Inhibition Assay | Inhibition of human SUP-T1 cell growth in a cell viability assay, IC50 = 0.26516 μM. | SANGER | |||
| MEG-01 | Growth Inhibition Assay | Inhibition of human MEG-01 cell growth in a cell viability assay, IC50 = 0.2687 μM. | SANGER | |||
| CHP-126 | Growth Inhibition Assay | Inhibition of human CHP-126 cell growth in a cell viability assay, IC50 = 0.26909 μM. | SANGER | |||
| HCC1599 | Growth Inhibition Assay | Inhibition of human HCC1599 cell growth in a cell viability assay, IC50 = 0.27841 μM. | SANGER | |||
| EB-3 | Growth Inhibition Assay | Inhibition of human EB-3 cell growth in a cell viability assay, IC50 = 0.29293 μM. | SANGER | |||
| WSU-NHL | Growth Inhibition Assay | Inhibition of human WSU-NHL cell growth in a cell viability assay, IC50 = 0.29672 μM. | SANGER | |||
| COLO-684 | Growth Inhibition Assay | Inhibition of human COLO-684 cell growth in a cell viability assay, IC50 = 0.3025 μM. | SANGER | |||
| LAMA-84 | Growth Inhibition Assay | Inhibition of human LAMA-84 cell growth in a cell viability assay, IC50 = 0.30816 μM. | SANGER | |||
| TGW | Growth Inhibition Assay | Inhibition of human TGW cell growth in a cell viability assay, IC50 = 0.31095 μM. | SANGER | |||
| NCI-SNU-1 | Growth Inhibition Assay | Inhibition of human NCI-SNU-1 cell growth in a cell viability assay, IC50 = 0.31106 μM. | SANGER | |||
| MHH-NB-11 | Growth Inhibition Assay | Inhibition of human MHH-NB-11 cell growth in a cell viability assay, IC50 = 0.32291 μM. | SANGER | |||
| COLO-824 | Growth Inhibition Assay | Inhibition of human COLO-824 cell growth in a cell viability assay, IC50 = 0.32838 μM. | SANGER | |||
| U-87-MG | Growth Inhibition Assay | Inhibition of human U-87-MG cell growth in a cell viability assay, IC50 = 0.33314 μM. | SANGER | |||
| HT | Growth Inhibition Assay | Inhibition of human HT cell growth in a cell viability assay, IC50 = 0.33482 μM. | SANGER | |||
| THP-1 | Growth Inhibition Assay | Inhibition of human THP-1 cell growth in a cell viability assay, IC50 = 0.34164 μM. | SANGER | |||
| HCC2218 | Growth Inhibition Assay | Inhibition of human HCC2218 cell growth in a cell viability assay, IC50 = 0.3418 μM. | SANGER | |||
| KARPAS-422 | Growth Inhibition Assay | Inhibition of human KARPAS-422 cell growth in a cell viability assay, IC50 = 0.34433 μM. | SANGER | |||
| EM-2 | Growth Inhibition Assay | Inhibition of human EM-2 cell growth in a cell viability assay, IC50 = 0.34443 μM. | SANGER | |||
| TUR | Growth Inhibition Assay | Inhibition of human TUR cell growth in a cell viability assay, IC50 = 0.34453 μM. | SANGER | |||
| MN-60 | Growth Inhibition Assay | Inhibition of human MN-60 cell growth in a cell viability assay, IC50 = 0.34596 μM. | SANGER | |||
| DMS-153 | Growth Inhibition Assay | Inhibition of human DMS-153 cell growth in a cell viability assay, IC50 = 0.37276 μM. | SANGER | |||
| J-RT3-T3-5 | Growth Inhibition Assay | Inhibition of human J-RT3-T3-5 cell growth in a cell viability assay, IC50 = 0.37831 μM. | SANGER | |||
| EVSA-T | Growth Inhibition Assay | Inhibition of human EVSA-T cell growth in a cell viability assay, IC50 = 0.38095 μM. | SANGER | |||
| SHP-77 | Growth Inhibition Assay | Inhibition of human SHP-77 cell growth in a cell viability assay, IC50 = 0.38298 μM. | SANGER | |||
| D-283MED | Growth Inhibition Assay | Inhibition of human D-283MED cell growth in a cell viability assay, IC50 = 0.38569 μM. | SANGER | |||
| GOTO | Growth Inhibition Assay | Inhibition of human GOTO cell growth in a cell viability assay, IC50 = 0.39536 μM. | SANGER | |||
| KMOE-2 | Growth Inhibition Assay | Inhibition of human KMOE-2 cell growth in a cell viability assay, IC50 = 0.39664 μM. | SANGER | |||
| U-266 | Growth Inhibition Assay | Inhibition of human U-266 cell growth in a cell viability assay, IC50 = 0.40452 μM. | SANGER | |||
| NCI-H226 | Growth Inhibition Assay | Inhibition of human NCI-H226 cell growth in a cell viability assay, IC50 = 0.40703 μM. | SANGER | |||
| KG-1 | Growth Inhibition Assay | Inhibition of human KG-1 cell growth in a cell viability assay, IC50 = 0.40876 μM. | SANGER | |||
| NCI-H1882 | Growth Inhibition Assay | Inhibition of human NCI-H1882 cell growth in a cell viability assay, IC50 = 0.40912 μM. | SANGER | |||
| KMS-12-PE | Growth Inhibition Assay | Inhibition of human KMS-12-PE cell growth in a cell viability assay, IC50 = 0.41877 μM. | SANGER | |||
| KM-H2 | Growth Inhibition Assay | Inhibition of human KM-H2 cell growth in a cell viability assay, IC50 = 0.42374 μM. | SANGER | |||
| NCI-H1417 | Growth Inhibition Assay | Inhibition of human NCI-H1417 cell growth in a cell viability assay, IC50 = 0.43916 μM. | SANGER | |||
| RPMI-8402 | Growth Inhibition Assay | Inhibition of human RPMI-8402 cell growth in a cell viability assay, IC50 = 0.44161 μM. | SANGER | |||
| EW-18 | Growth Inhibition Assay | Inhibition of human EW-18 cell growth in a cell viability assay, IC50 = 0.45104 μM. | SANGER | |||
| HH | Growth Inhibition Assay | Inhibition of human HH cell growth in a cell viability assay, IC50 = 0.45751 μM. | SANGER | |||
| NCI-H526 | Growth Inhibition Assay | Inhibition of human NCI-H526 cell growth in a cell viability assay, IC50 = 0.46847 μM. | SANGER | |||
| NEC8 | Growth Inhibition Assay | Inhibition of human NEC8 cell growth in a cell viability assay, IC50 = 0.48029 μM. | SANGER | |||
| P12-ICHIKAWA | Growth Inhibition Assay | Inhibition of human P12-ICHIKAWA cell growth in a cell viability assay, IC50 = 0.48862 μM. | SANGER | |||
| DB | Growth Inhibition Assay | Inhibition of human DB cell growth in a cell viability assay, IC50 = 0.48893 μM. | SANGER | |||
| SJRH30 | Growth Inhibition Assay | Inhibition of human SJRH30 cell growth in a cell viability assay, IC50 = 0.49478 μM. | SANGER | |||
| ATN-1 | Growth Inhibition Assay | Inhibition of human ATN-1 cell growth in a cell viability assay, IC50 = 0.49906 μM. | SANGER | |||
| ECC4 | Growth Inhibition Assay | Inhibition of human ECC4 cell growth in a cell viability assay, IC50 = 0.50901 μM. | SANGER | |||
| LU-65 | Growth Inhibition Assay | Inhibition of human LU-65 cell growth in a cell viability assay, IC50 = 0.52007 μM. | SANGER | |||
| NCI-H1092 | Growth Inhibition Assay | Inhibition of human NCI-H1092 cell growth in a cell viability assay, IC50 = 0.54078 μM. | SANGER | |||
| NCI-H187 | Growth Inhibition Assay | Inhibition of human NCI-H187 cell growth in a cell viability assay, IC50 = 0.54464 μM. | SANGER | |||
| LNCaP-Clone-FGC | Growth Inhibition Assay | Inhibition of human LNCaP-Clone-FGC cell growth in a cell viability assay, IC50 = 0.5472 μM. | SANGER | |||
| LC4-1 | Growth Inhibition Assay | Inhibition of human LC4-1 cell growth in a cell viability assay, IC50 = 0.56308 μM. | SANGER | |||
| NCI-H1436 | Growth Inhibition Assay | Inhibition of human NCI-H1436 cell growth in a cell viability assay, IC50 = 0.59043 μM. | SANGER | |||
| NCI-H2171 | Growth Inhibition Assay | Inhibition of human NCI-H2171 cell growth in a cell viability assay, IC50 = 0.62179 μM. | SANGER | |||
| LB647-SCLC | Growth Inhibition Assay | Inhibition of human LB647-SCLC cell growth in a cell viability assay, IC50 = 0.62241 μM. | SANGER | |||
| NCI-H2141 | Growth Inhibition Assay | Inhibition of human NCI-H2141 cell growth in a cell viability assay, IC50 = 0.63266 μM. | SANGER | |||
| NCI-H1304 | Growth Inhibition Assay | Inhibition of human NCI-H1304 cell growth in a cell viability assay, IC50 = 0.63388 μM. | SANGER | |||
| Raji | Growth Inhibition Assay | Inhibition of human Raji cell growth in a cell viability assay, IC50 = 0.66976 μM. | SANGER | |||
| NB69 | Growth Inhibition Assay | Inhibition of human NB69 cell growth in a cell viability assay, IC50 = 0.68575 μM. | SANGER | |||
| NCI-H510A | Growth Inhibition Assay | Inhibition of human NCI-H510A cell growth in a cell viability assay, IC50 = 0.69172 μM. | SANGER | |||
| COR-L88 | Growth Inhibition Assay | Inhibition of human COR-L88 cell growth in a cell viability assay, IC50 = 0.72456 μM. | SANGER | |||
| K052 | Growth Inhibition Assay | Inhibition of human K052 cell growth in a cell viability assay, IC50 = 0.79127 μM. | SANGER | |||
| SK-N-DZ | Growth Inhibition Assay | Inhibition of human SK-N-DZ cell growth in a cell viability assay, IC50 = 0.80231 μM. | SANGER | |||
| KY821 | Growth Inhibition Assay | Inhibition of human KY821 cell growth in a cell viability assay, IC50 = 0.81743 μM. | SANGER | |||
| MDA-MB-157 | Growth Inhibition Assay | Inhibition of human MDA-MB-157 cell growth in a cell viability assay, IC50 = 0.82406 μM. | SANGER | |||
| MS-1 | Growth Inhibition Assay | Inhibition of human MS-1 cell growth in a cell viability assay, IC50 = 0.82745 μM. | SANGER | |||
| L-540 | Growth Inhibition Assay | Inhibition of human L-540 cell growth in a cell viability assay, IC50 = 0.84843 μM. | SANGER | |||
| MDA-MB-361 | Growth Inhibition Assay | Inhibition of human MDA-MB-361 cell growth in a cell viability assay, IC50 = 0.86048 μM. | SANGER | |||
| NCI-SNU-5 | Growth Inhibition Assay | Inhibition of human NCI-SNU-5 cell growth in a cell viability assay, IC50 = 0.94375 μM. | SANGER | |||
| KARPAS-299 | Growth Inhibition Assay | Inhibition of human KARPAS-299 cell growth in a cell viability assay, IC50 = 0.94828 μM. | SANGER | |||
| NCI-H1694 | Growth Inhibition Assay | Inhibition of human NCI-H1694 cell growth in a cell viability assay, IC50 = 0.9773 μM. | SANGER | |||
| P30-OHK | Growth Inhibition Assay | Inhibition of human P30-OHK cell growth in a cell viability assay, IC50 = 0.99292 μM. | SANGER | |||
| LU-165 | Growth Inhibition Assay | Inhibition of human LU-165 cell growth in a cell viability assay, IC50 = 1.05606 μM. | SANGER | |||
| NB1 | Growth Inhibition Assay | Inhibition of human NB1 cell growth in a cell viability assay, IC50 = 1.11241 μM. | SANGER | |||
| UACC-812 | Growth Inhibition Assay | Inhibition of human UACC-812 cell growth in a cell viability assay, IC50 = 1.1488 μM. | SANGER | |||
| RL | Growth Inhibition Assay | Inhibition of human RL cell growth in a cell viability assay, IC50 = 1.16282 μM. | SANGER | |||
| NCI-H69 | Growth Inhibition Assay | Inhibition of human NCI-H69 cell growth in a cell viability assay, IC50 = 1.19391 μM. | SANGER | |||
| SW1417 | Growth Inhibition Assay | Inhibition of human SW1417 cell growth in a cell viability assay, IC50 = 1.23671 μM. | SANGER | |||
| NCI-H1155 | Growth Inhibition Assay | Inhibition of human NCI-H1155 cell growth in a cell viability assay, IC50 = 1.27588 μM. | SANGER | |||
| NCI-H1581 | Growth Inhibition Assay | Inhibition of human NCI-H1581 cell growth in a cell viability assay, IC50 = 1.44028 μM. | SANGER | |||
| MHH-CALL-2 | Growth Inhibition Assay | Inhibition of human MHH-CALL-2 cell growth in a cell viability assay, IC50 = 1.48514 μM. | SANGER | |||
| SCLC-21H | Growth Inhibition Assay | Inhibition of human SCLC-21H cell growth in a cell viability assay, IC50 = 1.67837 μM. | SANGER | |||
| LK-2 | Growth Inhibition Assay | Inhibition of human LK-2 cell growth in a cell viability assay, IC50 = 1.71089 μM. | SANGER | |||
| CPC-N | Growth Inhibition Assay | Inhibition of human CPC-N cell growth in a cell viability assay, IC50 = 1.76764 μM. | SANGER | |||
| SCC-3 | Growth Inhibition Assay | Inhibition of human SCC-3 cell growth in a cell viability assay, IC50 = 2.04311 μM. | SANGER | |||
| NCI-H446 | Growth Inhibition Assay | Inhibition of human NCI-H446 cell growth in a cell viability assay, IC50 = 2.05293 μM. | SANGER | |||
| RH-1 | Growth Inhibition Assay | Inhibition of human RH-1 cell growth in a cell viability assay, IC50 = 2.13933 μM. | SANGER | |||
| NCI-H345 | Growth Inhibition Assay | Inhibition of human NCI-H345 cell growth in a cell viability assay, IC50 = 2.14996 μM. | SANGER | |||
| A253 | Growth Inhibition Assay | Inhibition of human A253 cell growth in a cell viability assay, IC50 = 2.25377 μM. | SANGER | |||
| DMS-79 | Growth Inhibition Assay | Inhibition of human DMS-79 cell growth in a cell viability assay, IC50 = 2.36386 μM. | SANGER | |||
| SK-N-FI | Growth Inhibition Assay | Inhibition of human SK-N-FI cell growth in a cell viability assay, IC50 = 2.50034 μM. | SANGER | |||
| SK-MEL-1 | Growth Inhibition Assay | Inhibition of human SK-MEL-1 cell growth in a cell viability assay, IC50 = 2.61066 μM. | SANGER | |||
| HuO-3N1 | Growth Inhibition Assay | Inhibition of human HuO-3N1 cell growth in a cell viability assay, IC50 = 2.69128 μM. | SANGER | |||
| SIMA | Growth Inhibition Assay | Inhibition of human SIMA cell growth in a cell viability assay, IC50 = 2.72143 μM. | SANGER | |||
| SU-DHL-1 | Growth Inhibition Assay | Inhibition of human SU-DHL-1 cell growth in a cell viability assay, IC50 = 2.90246 μM. | SANGER | |||
| P31-FUJ | Growth Inhibition Assay | Inhibition of human P31-FUJ cell growth in a cell viability assay, IC50 = 3.01209 μM. | SANGER | |||
| A101D | Growth Inhibition Assay | Inhibition of human A101D cell growth in a cell viability assay, IC50 = 3.23245 μM. | SANGER | |||
| RPMI-8866 | Growth Inhibition Assay | Inhibition of human RPMI-8866 cell growth in a cell viability assay, IC50 = 3.58797 μM. | SANGER | |||
| NCI-H209 | Growth Inhibition Assay | Inhibition of human NCI-H209 cell growth in a cell viability assay, IC50 = 3.59247 μM. | SANGER | |||
| JVM-2 | Growth Inhibition Assay | Inhibition of human JVM-2 cell growth in a cell viability assay, IC50 = 3.62244 μM. | SANGER | |||
| LP-1 | Growth Inhibition Assay | Inhibition of human LP-1 cell growth in a cell viability assay, IC50 = 3.79042 μM. | SANGER | |||
| IST-SL1 | Growth Inhibition Assay | Inhibition of human IST-SL1 cell growth in a cell viability assay, IC50 = 3.91611 μM. | SANGER | |||
| TE-9 | Growth Inhibition Assay | Inhibition of human TE-9 cell growth in a cell viability assay, IC50 = 3.99619 μM. | SANGER | |||
| NCI-H1618 | Growth Inhibition Assay | Inhibition of human NCI-H1618 cell growth in a cell viability assay, IC50 = 4.17788 μM. | SANGER | |||
| COR-L105 | Growth Inhibition Assay | Inhibition of human COR-L105 cell growth in a cell viability assay, IC50 = 4.51462 μM. | SANGER | |||
| KU-19-19 | Growth Inhibition Assay | Inhibition of human KU-19-19 cell growth in a cell viability assay, IC50 = 4.56729 μM. | SANGER | |||
| HCC70 | Growth Inhibition Assay | Inhibition of human HCC70 cell growth in a cell viability assay, IC50 = 5.75167 μM. | SANGER | |||
| DG-75 | Growth Inhibition Assay | Inhibition of human DG-75 cell growth in a cell viability assay, IC50 = 6.18885 μM. | SANGER | |||
| CCF-STTG1 | Growth Inhibition Assay | Inhibition of human CCF-STTG1 cell growth in a cell viability assay, IC50 = 6.74902 μM. | SANGER | |||
| SBC-1 | Growth Inhibition Assay | Inhibition of human SBC-1 cell growth in a cell viability assay, IC50 = 6.88808 μM. | SANGER | |||
| CAL-85-1 | Growth Inhibition Assay | Inhibition of human CAL-85-1 cell growth in a cell viability assay, IC50 = 7.45587 μM. | SANGER | |||
| BxPC-3 | Growth Inhibition Assay | Inhibition of human BxPC-3 cell growth in a cell viability assay, IC50 = 7.56085 μM. | SANGER | |||
| YAPC | Growth Inhibition Assay | Inhibition of human YAPC cell growth in a cell viability assay, IC50 = 7.74286 μM. | SANGER | |||
| HCC1806 | Growth Inhibition Assay | Inhibition of human HCC1806 cell growth in a cell viability assay, IC50 = 8.04507 μM. | SANGER | |||
| SK-MEL-28 | Growth Inhibition Assay | Inhibition of human SK-MEL-28 cell growth in a cell viability assay, IC50 = 8.42441 μM. | SANGER | |||
| Ca9-22 | Growth Inhibition Assay | Inhibition of human Ca9-22 cell growth in a cell viability assay, IC50 = 8.62748 μM. | SANGER | |||
| NCI-H889 | Growth Inhibition Assay | Inhibition of human NCI-H889 cell growth in a cell viability assay, IC50 = 9.42457 μM. | SANGER | |||
| HT-3 | Growth Inhibition Assay | Inhibition of human HT-3 cell growth in a cell viability assay, IC50 = 9.4754 μM. | SANGER | |||
| H4 | Growth Inhibition Assay | Inhibition of human H4 cell growth in a cell viability assay, IC50 = 9.59864 μM. | SANGER | |||
| NCI-H2228 | Growth Inhibition Assay | Inhibition of human NCI-H2228 cell growth in a cell viability assay, IC50 = 9.87581 μM. | SANGER | |||
| M14 | Growth Inhibition Assay | Inhibition of human M14 cell growth in a cell viability assay, IC50 = 10.5224 μM. | SANGER | |||
| OVCAR-3 | Growth Inhibition Assay | Inhibition of human OVCAR-3 cell growth in a cell viability assay, IC50 = 11.4748 μM. | SANGER | |||
| HCC1187 | Growth Inhibition Assay | Inhibition of human HCC1187 cell growth in a cell viability assay, IC50 = 11.4776 μM. | SANGER | |||
| RT4 | Growth Inhibition Assay | Inhibition of human RT4 cell growth in a cell viability assay, IC50 = 11.6532 μM. | SANGER | |||
| SW962 | Growth Inhibition Assay | Inhibition of human SW962 cell growth in a cell viability assay, IC50 = 12.004 μM. | SANGER | |||
| BB49-HNC | Growth Inhibition Assay | Inhibition of human BB49-HNC cell growth in a cell viability assay, IC50 = 12.0436 μM. | SANGER | |||
| HCE-4 | Growth Inhibition Assay | Inhibition of human HCE-4 cell growth in a cell viability assay, IC50 = 12.7585 μM. | SANGER | |||
| KU812 | Growth Inhibition Assay | Inhibition of human KU812 cell growth in a cell viability assay, IC50 = 13.9562 μM. | SANGER | |||
| JAR | Growth Inhibition Assay | Inhibition of human JAR cell growth in a cell viability assay, IC50 = 14.113 μM. | SANGER | |||
| NCI-H2009 | Growth Inhibition Assay | Inhibition of human NCI-H2009 cell growth in a cell viability assay, IC50 = 14.287 μM. | SANGER | |||
| HuO9 | Growth Inhibition Assay | Inhibition of human HuO9 cell growth in a cell viability assay, IC50 = 14.4446 μM. | SANGER | |||
| U-118-MG | Growth Inhibition Assay | Inhibition of human U-118-MG cell growth in a cell viability assay, IC50 = 14.5013 μM. | SANGER | |||
| SH-4 | Growth Inhibition Assay | Inhibition of human SH-4 cell growth in a cell viability assay, IC50 = 14.942 μM. | SANGER | |||
| ES5 | Growth Inhibition Assay | Inhibition of human ES5 cell growth in a cell viability assay, IC50 = 15.3528 μM. | SANGER | |||
| NB6 | Growth Inhibition Assay | Inhibition of human NB6 cell growth in a cell viability assay, IC50 = 15.4452 μM. | SANGER | |||
| COLO-829 | Growth Inhibition Assay | Inhibition of human COLO-829 cell growth in a cell viability assay, IC50 = 15.6978 μM. | SANGER | |||
| NCI-H524 | Growth Inhibition Assay | Inhibition of human NCI-H524 cell growth in a cell viability assay, IC50 = 15.751 μM. | SANGER | |||
| HPAF-II | Growth Inhibition Assay | Inhibition of human HPAF-II cell growth in a cell viability assay, IC50 = 16.1017 μM. | SANGER | |||
| GB-1 | Growth Inhibition Assay | Inhibition of human GB-1 cell growth in a cell viability assay, IC50 = 16.5577 μM. | SANGER | |||
| LB771-HNC | Growth Inhibition Assay | Inhibition of human LB771-HNC cell growth in a cell viability assay, IC50 = 16.8228 μM. | SANGER | |||
| HuCCT1 | Growth Inhibition Assay | Inhibition of human HuCCT1 cell growth in a cell viability assay, IC50 = 17.176 μM. | SANGER | |||
| SW837 | Growth Inhibition Assay | Inhibition of human SW837 cell growth in a cell viability assay, IC50 = 17.3011 μM. | SANGER | |||
| NCI-N87 | Growth Inhibition Assay | Inhibition of human NCI-N87 cell growth in a cell viability assay, IC50 = 17.5302 μM. | SANGER | |||
| BEN | Growth Inhibition Assay | Inhibition of human BEN cell growth in a cell viability assay, IC50 = 17.7386 μM. | SANGER | |||
| Mewo | Growth Inhibition Assay | Inhibition of human Mewo cell growth in a cell viability assay, IC50 = 17.8382 μM. | SANGER | |||
| MPP-89 | Growth Inhibition Assay | Inhibition of human MPP-89 cell growth in a cell viability assay, IC50 = 18.2784 μM. | SANGER | |||
| NOS-1 | Growth Inhibition Assay | Inhibition of human NOS-1 cell growth in a cell viability assay, IC50 = 18.3793 μM. | SANGER | |||
| Ca-Ski | Growth Inhibition Assay | Inhibition of human Ca-Ski cell growth in a cell viability assay, IC50 = 19.7889 μM. | SANGER | |||
| SW1088 | Growth Inhibition Assay | Inhibition of human SW1088 cell growth in a cell viability assay, IC50 = 19.9447 μM. | SANGER | |||
| Capan-2 | Growth Inhibition Assay | Inhibition of human Capan-2 cell growth in a cell viability assay, IC50 = 20.3327 μM. | SANGER | |||
| A498 | Growth Inhibition Assay | Inhibition of human A498 cell growth in a cell viability assay, IC50 = 20.4092 μM. | SANGER | |||
| COLO-800 | Growth Inhibition Assay | Inhibition of human COLO-800 cell growth in a cell viability assay, IC50 = 20.9232 μM. | SANGER | |||
| OVCAR-4 | Growth Inhibition Assay | Inhibition of human OVCAR-4 cell growth in a cell viability assay, IC50 = 21.2508 μM. | SANGER | |||
| K-562 | Growth Inhibition Assay | Inhibition of human K-562 cell growth in a cell viability assay, IC50 = 21.9049 μM. | SANGER | |||
| IA-LM | Growth Inhibition Assay | Inhibition of human IA-LM cell growth in a cell viability assay, IC50 = 22.3771 μM. | SANGER | |||
| SK-LMS-1 | Growth Inhibition Assay | Inhibition of human SK-LMS-1 cell growth in a cell viability assay, IC50 = 22.4057 μM. | SANGER | |||
| CAL-51 | Growth Inhibition Assay | Inhibition of human CAL-51 cell growth in a cell viability assay, IC50 = 23.0872 μM. | SANGER | |||
| NCI-H1651 | Growth Inhibition Assay | Inhibition of human NCI-H1651 cell growth in a cell viability assay, IC50 = 24.1303 μM. | SANGER | |||
| GMS-10 | Growth Inhibition Assay | Inhibition of human GMS-10 cell growth in a cell viability assay, IC50 = 24.4376 μM. | SANGER | |||
| NCI-H2347 | Growth Inhibition Assay | Inhibition of human NCI-H2347 cell growth in a cell viability assay, IC50 = 24.6005 μM. | SANGER | |||
| MEL-JUSO | Growth Inhibition Assay | Inhibition of human MEL-JUSO cell growth in a cell viability assay, IC50 = 24.6906 μM. | SANGER | |||
| SW948 | Growth Inhibition Assay | Inhibition of human SW948 cell growth in a cell viability assay, IC50 = 24.9296 μM. | SANGER | |||
| MKN45 | Growth Inhibition Assay | Inhibition of human MKN45 cell growth in a cell viability assay, IC50 = 26.4465 μM. | SANGER | |||
| HCC1954 | Growth Inhibition Assay | Inhibition of human HCC1954 cell growth in a cell viability assay, IC50 = 26.4502 μM. | SANGER | |||
| SAS | Growth Inhibition Assay | Inhibition of human SAS cell growth in a cell viability assay, IC50 = 26.4732 μM. | SANGER | |||
| SCC-25 | Growth Inhibition Assay | Inhibition of human SCC-25 cell growth in a cell viability assay, IC50 = 26.5693 μM. | SANGER | |||
| MKN28 | Growth Inhibition Assay | Inhibition of human MKN28 cell growth in a cell viability assay, IC50 = 26.7093 μM. | SANGER | |||
| T84 | Growth Inhibition Assay | Inhibition of human T84 cell growth in a cell viability assay, IC50 = 26.7743 μM. | SANGER | |||
| HCT-15 | Growth Inhibition Assay | Inhibition of human HCT-15 cell growth in a cell viability assay, IC50 = 26.9389 μM. | SANGER | |||
| DOK | Growth Inhibition Assay | Inhibition of human DOK cell growth in a cell viability assay, IC50 = 27.0723 μM. | SANGER | |||
| SK-MEL-24 | Growth Inhibition Assay | Inhibition of human SK-MEL-24 cell growth in a cell viability assay, IC50 = 28.0766 μM. | SANGER | |||
| HCC1143 | Growth Inhibition Assay | Inhibition of human HCC1143 cell growth in a cell viability assay, IC50 = 28.2552 μM. | SANGER | |||
| JEG-3 | Growth Inhibition Assay | Inhibition of human JEG-3 cell growth in a cell viability assay, IC50 = 28.4578 μM. | SANGER | |||
| SF539 | Growth Inhibition Assay | Inhibition of human SF539 cell growth in a cell viability assay, IC50 = 28.6049 μM. | SANGER | |||
| NB12 | Growth Inhibition Assay | Inhibition of human NB12 cell growth in a cell viability assay, IC50 = 28.8033 μM. | SANGER | |||
| Hs-578-T | Growth Inhibition Assay | Inhibition of human Hs-578-T cell growth in a cell viability assay, IC50 = 29.1197 μM. | SANGER | |||
| HCT-116 | Growth Inhibition Assay | Inhibition of human HCT-116 cell growth in a cell viability assay, IC50 = 29.4423 μM. | SANGER | |||
| EW-3 | Growth Inhibition Assay | Inhibition of human EW-3 cell growth in a cell viability assay, IC50 = 29.5269 μM. | SANGER | |||
| 639-V | Growth Inhibition Assay | Inhibition of human 639-V cell growth in a cell viability assay, IC50 = 29.8546 μM. | SANGER | |||
| IGR-1 | Growth Inhibition Assay | Inhibition of human IGR-1 cell growth in a cell viability assay, IC50 = 30.0191 μM. | SANGER | |||
| LS-411N | Growth Inhibition Assay | Inhibition of human LS-411N cell growth in a cell viability assay, IC50 = 30.351 μM. | SANGER | |||
| D-502MG | Growth Inhibition Assay | Inhibition of human D-502MG cell growth in a cell viability assay, IC50 = 30.3725 μM. | SANGER | |||
| AN3-CA | Growth Inhibition Assay | Inhibition of human AN3-CA cell growth in a cell viability assay, IC50 = 31.4329 μM. | SANGER | |||
| RCC10RGB | Growth Inhibition Assay | Inhibition of human RCC10RGB cell growth in a cell viability assay, IC50 = 31.4464 μM. | SANGER | |||
| GT3TKB | Growth Inhibition Assay | Inhibition of human GT3TKB cell growth in a cell viability assay, IC50 = 31.7096 μM. | SANGER | |||
| BB30-HNC | Growth Inhibition Assay | Inhibition of human BB30-HNC cell growth in a cell viability assay, IC50 = 32.5217 μM. | SANGER | |||
| HT-144 | Growth Inhibition Assay | Inhibition of human HT-144 cell growth in a cell viability assay, IC50 = 32.7065 μM. | SANGER | |||
| NCI-H1355 | Growth Inhibition Assay | Inhibition of human NCI-H1355 cell growth in a cell viability assay, IC50 = 33.3495 μM. | SANGER | |||
| D-542MG | Growth Inhibition Assay | Inhibition of human D-542MG cell growth in a cell viability assay, IC50 = 33.3722 μM. | SANGER | |||
| NCI-H727 | Growth Inhibition Assay | Inhibition of human NCI-H727 cell growth in a cell viability assay, IC50 = 33.4114 μM. | SANGER | |||
| NCI-H1755 | Growth Inhibition Assay | Inhibition of human NCI-H1755 cell growth in a cell viability assay, IC50 = 33.5236 μM. | SANGER | |||
| KS-1 | Growth Inhibition Assay | Inhibition of human KS-1 cell growth in a cell viability assay, IC50 = 33.6298 μM. | SANGER | |||
| SW626 | Growth Inhibition Assay | Inhibition of human SW626 cell growth in a cell viability assay, IC50 = 33.7643 μM. | SANGER | |||
| MZ1-PC | Growth Inhibition Assay | Inhibition of human MZ1-PC cell growth in a cell viability assay, IC50 = 33.8103 μM. | SANGER | |||
| NCI-H1650 | Growth Inhibition Assay | Inhibition of human NCI-H1650 cell growth in a cell viability assay, IC50 = 34.326 μM. | SANGER | |||
| LCLC-97TM1 | Growth Inhibition Assay | Inhibition of human LCLC-97TM1 cell growth in a cell viability assay, IC50 = 34.8526 μM. | SANGER | |||
| HTC-C3 | Growth Inhibition Assay | Inhibition of human HTC-C3 cell growth in a cell viability assay, IC50 = 35.022 μM. | SANGER | |||
| SNU-387 | Growth Inhibition Assay | Inhibition of human SNU-387 cell growth in a cell viability assay, IC50 = 35.2684 μM. | SANGER | |||
| IM-9 | Growth Inhibition Assay | Inhibition of human IM-9 cell growth in a cell viability assay, IC50 = 35.5165 μM. | SANGER | |||
| KYSE-180 | Growth Inhibition Assay | Inhibition of human KYSE-180 cell growth in a cell viability assay, IC50 = 35.6521 μM. | SANGER | |||
| HuH-7 | Growth Inhibition Assay | Inhibition of human HuH-7 cell growth in a cell viability assay, IC50 = 35.6756 μM. | SANGER | |||
| CP66-MEL | Growth Inhibition Assay | Inhibition of human CP66-MEL cell growth in a cell viability assay, IC50 = 35.7968 μM. | SANGER | |||
| 22RV1 | Growth Inhibition Assay | Inhibition of human 22RV1 cell growth in a cell viability assay, IC50 = 35.8423 μM. | SANGER | |||
| SW48 | Growth Inhibition Assay | Inhibition of human SW48 cell growth in a cell viability assay, IC50 = 36.2464 μM. | SANGER | |||
| COLO-205 | Growth Inhibition Assay | Inhibition of human COLO-205 cell growth in a cell viability assay, IC50 = 37.2589 μM. | SANGER | |||
| MFH-ino | Growth Inhibition Assay | Inhibition of human MFH-ino cell growth in a cell viability assay, IC50 = 37.3918 μM. | SANGER | |||
| ECC12 | Growth Inhibition Assay | Inhibition of human ECC12 cell growth in a cell viability assay, IC50 = 37.7142 μM. | SANGER | |||
| CAL-72 | Growth Inhibition Assay | Inhibition of human CAL-72 cell growth in a cell viability assay, IC50 = 37.7221 μM. | SANGER | |||
| KNS-42 | Growth Inhibition Assay | Inhibition of human KNS-42 cell growth in a cell viability assay, IC50 = 37.8215 μM. | SANGER | |||
| U031 | Growth Inhibition Assay | Inhibition of human U031 cell growth in a cell viability assay, IC50 = 37.827 μM. | SANGER | |||
| EFE-184 | Growth Inhibition Assay | Inhibition of human EFE-184 cell growth in a cell viability assay, IC50 = 37.8863 μM. | SANGER | |||
| TCCSUP | Growth Inhibition Assay | Inhibition of human TCCSUP cell growth in a cell viability assay, IC50 = 38.3341 μM. | SANGER | |||
| TE-11 | Growth Inhibition Assay | Inhibition of human TE-11 cell growth in a cell viability assay, IC50 = 39.2614 μM. | SANGER | |||
| SK-MEL-3 | Growth Inhibition Assay | Inhibition of human SK-MEL-3 cell growth in a cell viability assay, IC50 = 39.5577 μM. | SANGER | |||
| NCI-H1703 | Growth Inhibition Assay | Inhibition of human NCI-H1703 cell growth in a cell viability assay, IC50 = 39.5618 μM. | SANGER | |||
| NCI-H748 | Growth Inhibition Assay | Inhibition of human NCI-H748 cell growth in a cell viability assay, IC50 = 39.6102 μM. | SANGER | |||
| HuP-T3 | Growth Inhibition Assay | Inhibition of human HuP-T3 cell growth in a cell viability assay, IC50 = 39.8492 μM. | SANGER | |||
| GCT | Growth Inhibition Assay | Inhibition of human GCT cell growth in a cell viability assay, IC50 = 40.2935 μM. | SANGER | |||
| LU-99A | Growth Inhibition Assay | Inhibition of human LU-99A cell growth in a cell viability assay, IC50 = 40.6473 μM. | SANGER | |||
| TGBC24TKB | Growth Inhibition Assay | Inhibition of human TGBC24TKB cell growth in a cell viability assay, IC50 = 40.7998 μM. | SANGER | |||
| NCI-H1993 | Growth Inhibition Assay | Inhibition of human NCI-H1993 cell growth in a cell viability assay, IC50 = 41.1086 μM. | SANGER | |||
| 769-P | Growth Inhibition Assay | Inhibition of human 769-P cell growth in a cell viability assay, IC50 = 41.226 μM. | SANGER | |||
| NCI-H2126 | Growth Inhibition Assay | Inhibition of human NCI-H2126 cell growth in a cell viability assay, IC50 = 41.3099 μM. | SANGER | |||
| NCI-H1563 | Growth Inhibition Assay | Inhibition of human NCI-H1563 cell growth in a cell viability assay, IC50 = 41.353 μM. | SANGER | |||
| NCI-H82 | Growth Inhibition Assay | Inhibition of human NCI-H82 cell growth in a cell viability assay, IC50 = 41.4039 μM. | SANGER | |||
| SW1783 | Growth Inhibition Assay | Inhibition of human SW1783 cell growth in a cell viability assay, IC50 = 41.6769 μM. | SANGER | |||
| NCI-H1648 | Growth Inhibition Assay | Inhibition of human NCI-H1648 cell growth in a cell viability assay, IC50 = 41.9392 μM. | SANGER | |||
| A388 | Growth Inhibition Assay | Inhibition of human A388 cell growth in a cell viability assay, IC50 = 42.1236 μM. | SANGER | |||
| SK-MEL-30 | Growth Inhibition Assay | Inhibition of human SK-MEL-30 cell growth in a cell viability assay, IC50 = 42.3151 μM. | SANGER | |||
| EKVX | Growth Inhibition Assay | Inhibition of human EKVX cell growth in a cell viability assay, IC50 = 42.3726 μM. | SANGER | |||
| A172 | Growth Inhibition Assay | Inhibition of human A172 cell growth in a cell viability assay, IC50 = 42.8624 μM. | SANGER | |||
| OMC-1 | Growth Inhibition Assay | Inhibition of human OMC-1 cell growth in a cell viability assay, IC50 = 42.9399 μM. | SANGER | |||
| PANC-03-27 | Growth Inhibition Assay | Inhibition of human PANC-03-27 cell growth in a cell viability assay, IC50 = 43.157 μM. | SANGER | |||
| NCI-H747 | Growth Inhibition Assay | Inhibition of human NCI-H747 cell growth in a cell viability assay, IC50 = 43.3223 μM. | SANGER | |||
| HT55 | Growth Inhibition Assay | Inhibition of human HT55 cell growth in a cell viability assay, IC50 = 43.7863 μM. | SANGER | |||
| BV-173 | Growth Inhibition Assay | Inhibition of human BV-173 cell growth in a cell viability assay, IC50 = 43.7946 μM. | SANGER | |||
| LS-1034 | Growth Inhibition Assay | Inhibition of human LS-1034 cell growth in a cell viability assay, IC50 = 43.9899 μM. | SANGER | |||
| NUGC-3 | Growth Inhibition Assay | Inhibition of human NUGC-3 cell growth in a cell viability assay, IC50 = 44.0773 μM. | SANGER | |||
| IST-MEL1 | Growth Inhibition Assay | Inhibition of human IST-MEL1 cell growth in a cell viability assay, IC50 = 44.2633 μM. | SANGER | |||
| 8305C | Growth Inhibition Assay | Inhibition of human 8305C cell growth in a cell viability assay, IC50 = 44.31 μM. | SANGER | |||
| IMR-5 | Growth Inhibition Assay | Inhibition of human IMR-5 cell growth in a cell viability assay, IC50 = 44.5743 μM. | SANGER | |||
| TE-6 | Growth Inhibition Assay | Inhibition of human TE-6 cell growth in a cell viability assay, IC50 = 44.8539 μM. | SANGER | |||
| MZ2-MEL | Growth Inhibition Assay | Inhibition of human MZ2-MEL cell growth in a cell viability assay, IC50 = 45.0112 μM. | SANGER | |||
| RT-112 | Growth Inhibition Assay | Inhibition of human RT-112 cell growth in a cell viability assay, IC50 = 45.4472 μM. | SANGER | |||
| SCC-9 | Growth Inhibition Assay | Inhibition of human SCC-9 cell growth in a cell viability assay, IC50 = 45.5616 μM. | SANGER | |||
| SCC-4 | Growth Inhibition Assay | Inhibition of human SCC-4 cell growth in a cell viability assay, IC50 = 45.5626 μM. | SANGER | |||
| CAL-54 | Growth Inhibition Assay | Inhibition of human CAL-54 cell growth in a cell viability assay, IC50 = 45.6635 μM. | SANGER | |||
| LoVo | Growth Inhibition Assay | Inhibition of human LoVo cell growth in a cell viability assay, IC50 = 45.6839 μM. | SANGER | |||
| NCI-H358 | Growth Inhibition Assay | Inhibition of human NCI-H358 cell growth in a cell viability assay, IC50 = 45.7286 μM. | SANGER | |||
| K5 | Growth Inhibition Assay | Inhibition of human K5 cell growth in a cell viability assay, IC50 = 46.029 μM. | SANGER | |||
| LB1047-RCC | Growth Inhibition Assay | Inhibition of human LB1047-RCC cell growth in a cell viability assay, IC50 = 46.4256 μM. | SANGER | |||
| MES-SA | Growth Inhibition Assay | Inhibition of human MES-SA cell growth in a cell viability assay, IC50 = 46.5121 μM. | SANGER | |||
| UACC-257 | Growth Inhibition Assay | Inhibition of human UACC-257 cell growth in a cell viability assay, IC50 = 46.5175 μM. | SANGER | |||
| NCI-H2342 | Growth Inhibition Assay | Inhibition of human NCI-H2342 cell growth in a cell viability assay, IC50 = 46.8809 μM. | SANGER | |||
| TE-15 | Growth Inhibition Assay | Inhibition of human TE-15 cell growth in a cell viability assay, IC50 = 46.8909 μM. | SANGER | |||
| SK-MES-1 | Growth Inhibition Assay | Inhibition of human SK-MES-1 cell growth in a cell viability assay, IC50 = 47.3821 μM. | SANGER | |||
| NCI-H322M | Growth Inhibition Assay | Inhibition of human NCI-H322M cell growth in a cell viability assay, IC50 = 47.6561 μM. | SANGER | |||
| SK-OV-3 | Growth Inhibition Assay | Inhibition of human SK-OV-3 cell growth in a cell viability assay, IC50 = 47.7452 μM. | SANGER | |||
| PANC-08-13 | Growth Inhibition Assay | Inhibition of human PANC-08-13 cell growth in a cell viability assay, IC50 = 47.9641 μM. | SANGER | |||
| 786-0 | Growth Inhibition Assay | Inhibition of human 786-0 cell growth in a cell viability assay, IC50 = 48.073 μM. | SANGER | |||
| NCI-H460 | Growth Inhibition Assay | Inhibition of human NCI-H460 cell growth in a cell viability assay, IC50 = 48.1255 μM. | SANGER | |||
| D-336MG | Growth Inhibition Assay | Inhibition of human D-336MG cell growth in a cell viability assay, IC50 = 48.4845 μM. | SANGER | |||
| SNG-M | Growth Inhibition Assay | Inhibition of human SNG-M cell growth in a cell viability assay, IC50 = 48.6701 μM. | SANGER | |||
| SK-CO-1 | Growth Inhibition Assay | Inhibition of human SK-CO-1 cell growth in a cell viability assay, IC50 = 49.0025 μM. | SANGER | |||
| CAL-27 | Growth Inhibition Assay | Inhibition of human CAL-27 cell growth in a cell viability assay, IC50 = 49.5334 μM. | SANGER | |||
| 8505C | Growth Inhibition Assay | Inhibition of human 8505C cell growth in a cell viability assay, IC50 = 49.5364 μM. | SANGER | |||
| MCF7 | Growth Inhibition Assay | Inhibition of human MCF7 cell growth in a cell viability assay, IC50 = 49.5865 μM. | SANGER | |||
| T47D | Growth Inhibition Assay | Inhibition of human T47D cell growth in a cell viability assay, IC50 = 49.7007 μM. | SANGER | |||
| YKG-1 | Growth Inhibition Assay | Inhibition of human YKG-1 cell growth in a cell viability assay, IC50 = 49.9412 μM. | SANGER | |||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 100 mg/mL
(277.47 mM)
Warmed with 50°C water bath;
Ultrasonicated;
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
Dilution Calculator
Molecular Weight Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 360.39 | Formula | C20H12N2O3S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 844499-71-4 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | C1=CC=C(C(=C1)C2=CC=C(C=C2)C3=CSC4=C3C(=C(C(=O)N4)C#N)O)O | ||
Mechanism of Action
| Targets/IC50/Ki |
AMPK
(Cell-free assay) 0.8 μM(EC50)
Fatty acid synthesis
(in primary rat hepatocytes) 3.2 μM
|
|---|---|
| In vitro |
A-769662 stimulates partially purified rat liver AMPK with EC50 with 0.8 μM. This compound activates AMPK purified from multiple tissues and species in a dose-responsive manner with modest variations in observed EC50s. EC50s determined for this chemical using partially purified AMPK extracts from rat heart, rat muscle, or human embryonic kidney cells (HEKs) are 2.2 mM, 1.9 mM, or 1.1 mM, respectively. A 4 hours treatment of primary rat hepatocytes with this compound dose-dependently increases ACC phosphorylation, which correlated inhibition of fatty acid synthesis with IC50 of 3.2 μM. It also inhibits fatty acid sythesis in mouse hepatocytes with IC50 with 3.6 μM This compound activates AMPK both allosterically and by inhibiting dephosphorylation of AMPK on Thr-172, similar to the effects of AMP. It inhibits proteasomal function by an AMPK-independent mechanism. This chemical affects the in vitro activity of purified 26S proteasomes but not the in vitro activity of purified 20S proteasomes. It has toxic effects on MEF cells. A recent research shows this compound inhibited cell proliferation and DNA synthesis. |
| Kinase Assay |
96-well AMPK assay
|
|
AMPK activity is measured by monitoring phosphorylation of the SAMS peptide substrate (20 μM in standard assays and 100 μM in additivity assays) following a previously described protocol (Anderson et al., 2004). To determine whether A-769662-induced AMPK activation occurs in a reversible manner, AMP or this compound are preincubated with rat liver AMPK for 10 minutes at 20 times standard assay concentrations prior to dilution and measurement of AMPK activity.
|
|
| In vivo |
Short-term treatment of normal Sprague Dawley rats with A-769662 decreases liver malonyl CoA levels and the respiratory exchange ratio, VCO2/VO2, indicating an increased rate of whole-body fatty acid oxidation. Treatment of ob/ob mice with 30 mg/kg b.i.d. of this compound decreases hepatic expression of PEPCK, G6Pase, and FAS, lowers plasma glucose by 40%, reduced body weight gain and significantly decreases both plasma and liver triglyceride levels. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | pCofilin / Cofilin / detyrosinated alpha tubulin / acetylated alpha tubulin p-AMPK / AMPK / p-S6K / S6K / p-ACC |
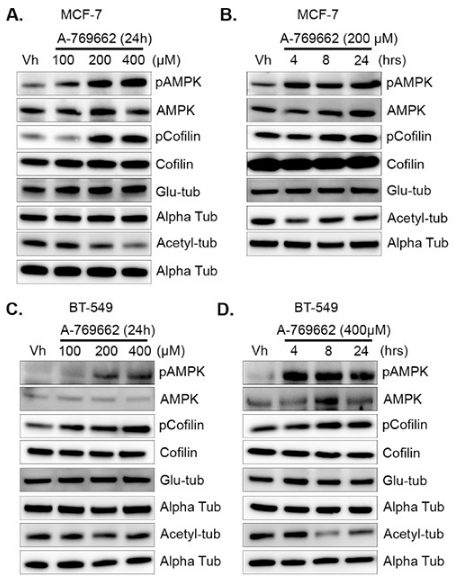
|
26431377 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































