research use only
SGI-1027 DNA Methyltransferase inhibitor
Cat.No.S7276
Chemical Structure
Molecular Weight: 461.52
Quality Control
| Related Targets | HDAC JAK BET Histone Methyltransferase PKC PARP HIF PRMT EZH2 AMPK |
|---|---|
| Other DNA Methyltransferase Inhibitors | RG108 Zebularine (NSC 309132) GSK3685032 Gamma-Oryzanol CM272 Bobcat339 β-thujaplicin GSK-3484862 DC-05 2'-Deoxy-5-Fluorocytidine |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human U937 cells | Cytotoxicity assay | 48 h | Cytotoxicity against human U937 cells after 48 hrs by trypan blue exclusion assay, IC50=1.7 μM | |||
| human KARPAS299 cells | Proliferation assay | 2-4 days | Antiproliferative activity against human KARPAS299 cells after 2 to 4 days by ATPlite 1step luminescence assay, EC50=1.8 μM | |||
| human KG1 cells | Proliferation assay | 2-4 days | Antiproliferative activity against human KG1 cells after 2 to 4 days by ATPlite 1step luminescence assay, EC50=4.4 μM | |||
| human MDA-MB-231 cells | Cytotoxicity assay | 48 h | Cytotoxicity against human MDA-MB-231 cells after 48 hrs by trypan blue exclusion assay, IC50=4.8 μM | |||
| human PC3 cells | Cytotoxicity assay | 48 h | Cytotoxicity against human PC3 cells after 48 hrs by trypan blue exclusion assay, IC50=6.5 μM | |||
| human Raji cells | Cytotoxicity assay | 48 h | Cytotoxicity against human Raji cells after 48 hrs by trypan blue exclusion assay, IC50=9.1 μM | |||
| human PBMC | Cytotoxicity assay | 48 h | Cytotoxicity against human PBMC after 48 hrs by trypan blue exclusion assay, IC50=23.8 μM | |||
| human HCT116 cells | Cytotoxicity assay | Cytotoxicity against human HCT116 cells assessed as viability, TD50=25 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 68 mg/mL
(147.33 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 461.52 | Formula | C27H23N7O |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1020149-73-8 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | DNA Methyltransferase Inhibitor II | Smiles | CC1=CC(=NC(=N1)N)NC2=CC=C(C=C2)NC(=O)C3=CC=C(C=C3)NC4=CC=NC5=CC=CC=C54 | ||
Mechanism of Action
| Features |
Potential for use in epigenetic cancer therapy.
|
|---|---|
| Targets/IC50/Ki |
DNMT1
(Cell-free assay) 6 μM
DNMT3B
(Cell-free assay) 7.5 μM
DNMT3A
(Cell-free assay) 8 μM
|
| In vitro |
SGI-1027 inhibits DNA methylation by directly inhibiting DNMTs, and results in selective degradation of DNMT1 in a wide variety of human cancer cell lines. This compound exhibits minimal or no cytotoxic effect in rat hepatoma H4IIE cells. This chemical (0-100 μM) exhibits a moderate pro-apoptotic effect on U937 human leukemia cell line with no relevant changes on the cell cycle. |
| Kinase Assay |
DNA methyltransferase (CpG methyltransferase) assay
|
|
DNA methylase activity is assayed by measuring the incorporation of 3H1-methyl group from Ado-Met into DNA using DE-81 ion exchange filter binding assay with some modifications. Human recombinant DNMT1, recombinant mouse Dnmt3a/ Dnmt3b (500 ng) is incubated with 500 ng of poly(dI-dC) or hemimethylated DNA duplex and 75 or 150 nM (0.275μCi or 0.55μCi) of [methyl-3H]-Sadenosylmethionine (Ado-Met) in a total volume of 50 μl at 37°C for 1hr. or M. Sss I is assayed in the supplier’s buffer. Each reaction is performed in duplicate and included controls with no inhibitor or no DNA. The reaction is stopped by soaking reaction mixture onto a Whatman DE-81 ion exchange filter disc, washed (five times, 10 min each, with 0.5M Na-phosphate buffer; pH 7.0) dried and counted in a scintillation counter. The background radioactivity (no DNA control) is subtracted from the values obtained with reaction mixtures containing DNA and the radioactivity obtained in the reaction without any inhibitor is considered as 100% activity. IC50 is determined by interpolation from the plot of percent activity versus inhibitor concentration. To determine the nature of inhibition of DNMTase activity by this compound, DNMT1 enzyme activityis measured in presence of a fixed concentration of this chemical (0, 2.5, 5, and 10μM) while one of the two (Ado-Met or DNA) was varied in a particular reaction mixture. At a fixed concentration of DNA (500 ng) varying concentrations of Ado-Met used are from 25-500 nM, respectively. Similarly, final DNA concentrations are varied from (25-500ng) at 75 nM Ado-Met.
|
|
| In vivo |
SGI-1027 (DNA Methyltransferase Inhibitor II) is a DNMT inhibitor . |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
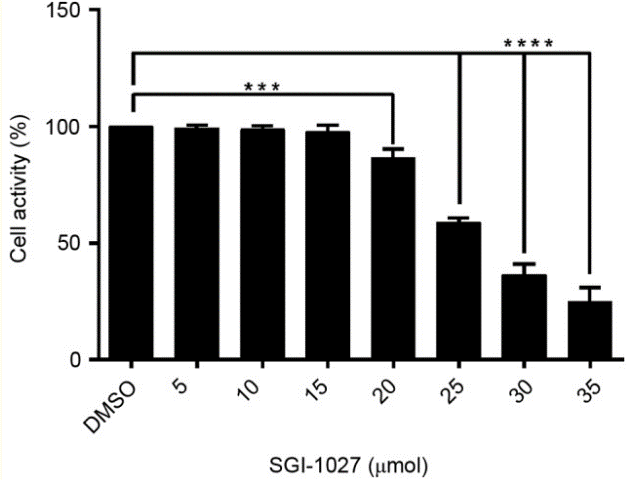
| Growth inhibition assay | Cell viability |
|
30344731 |
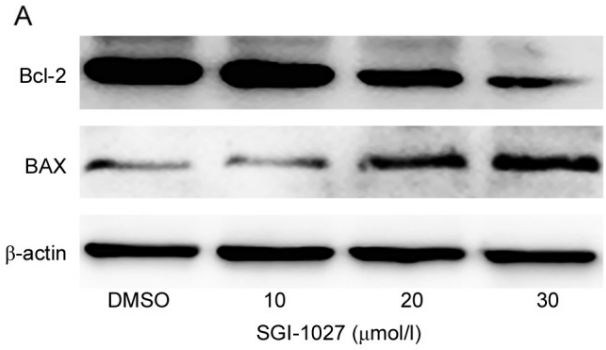
| Western blot | Bcl-2 / Bax DNMT1 / TIMP3 / P16 |
|
30344731 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































