research use only
Avutometinib (Ro5126766, CH5126766) RAF/MEK Inhibitor
Cat.No.S7170
Chemical Structure
Molecular Weight: 471.46
Quality Control
| Related Targets | ERK p38 MAPK JNK MEK Ras KRas S6 Kinase MAP4K TAK1 Mixed Lineage Kinase |
|---|---|
| Other Raf Inhibitors | LY3009120 Exarafenib (KIN-2787) GDC-0879 PLX-4720 AZ 628 SB590885 TAK-632 GW5074 RAF265 (CHIR-265) PLX8394 (Plixorafenib, FORE8394) |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| HCT116 cell | Function assay | 96 h | Inhibition of human HCT116 cell growth after 96 hrs by counting kit-8 analysis, IC50=0.04 μM | 24900832 | ||
| human C32 cells | Growth inhibition assay | Growth inhibition of human C32 cells harboring R-Raf V600E mutant, IC50=0.047 μM | 24900832 | |||
| human PANC1 cells | Function assay | 10 μM | 1 h | Inhibition of MEK1 in human PANC1 cells assessed as reduction in pErk1/2 level at 10 uM after 1 hr by Western blotting analysis | 25766633 | |
| human A549 cells | Function assay | 10 μM | 1 h | Inhibition of MEK1 in human A549 cells assessed as reduction in pErk1/2 level at 10 uM after 1 hr by Western blotting analysis | 25766633 | |
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 94 mg/mL
(199.38 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 471.46 | Formula | C21H18FN5O5S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 946128-88-7 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | RO5126766,CH5126766,VS 6766, CKI-27, R-7304, RG-7304 | Smiles | CC1=C(C(=O)OC2=C1C=CC(=C2)OC3=NC=CC=N3)CC4=C(C(=NC=C4)NS(=O)(=O)NC)F | ||
Mechanism of Action
| Targets/IC50/Ki |
BRAF V600E
(cell-free assay) 8.2 nM
BRAF
(cell-free assay) 19 nM
CRAF
(cell-free assay) 56 nM
MEK1
(cell-free assay) 160 nM
|
|---|---|
| In vitro |
In HCT116 KRAS-mutant colorectal cancer cells, CH5126766 significantly reduces the levels of phospho-MEK and phospho-ERK. CH5126766 inhibits RAF kinase by binding to MEK1, and causes MEK to become a dominant negative inhibitor of RAF. In Raf or RAS-mutant cell lines SK-MEL-28, SK-MEL-2, MIAPaCa-2, SW480, HCT116, and PC3 cells, CH5126766 inhibits cell growth with IC50 of 65, 28, 40, 46, and 277 nM, respectively. In two melanoma cell lines with the BRAF V600E or NRAS mutation, RO5126766 induces G1 cell cycle arrest accompanied by up-regulation of the CDK inhibitor p27 and down-regulation of cyclinD1. |
| Kinase Assay |
MEK and RAF kinase enzyme assays
|
|
The inhibitory activities against CRAF, BRAF, or BRAF V600E enzymes are measured by quantification of phosphorylation of inactive K97R MEK1 [MEK1] by recombinant RAF proteins [BRAF: B-RAF wt, BRAF V600E: B-RAF V600E or CRAF: Raf-1] with Europium-anti-MEK1/2 (pSer218/222) antibody and SureLight allophycocyanine-anti-6his antibody by measuring time-resolved fluorescence (TRF). Inhibition of MEK1 is evaluated by a coupled assay with active MEK1 (MEK1 S218E/S222E) and unactive dephosphorylated ERK2 (MAP kinase 2/Erk 2). The phosphorylation of a fluorescent-labeled peptide substrate (FAM-Erktide, IPTTPITTTYFFFK-5FAM-COOH) by ERK2 is quantified by using the IMAP FP Screening Express Kit.
|
|
| In vivo |
In an HCT116 (G13D KRAS) mouse xenograft model, CH5126766 (25 mg/kg, p.o.) inhibits ERK signaling output more effectively than a standard MEK inhibitor that induces MEK phosphorylation and has potent antitumor activity. In the HCT116 (K-ras) and COLO205 (B-raf) mutant xenografts, CH5126766 (0.3 mg/kg) causes significant decreases in [18 F]FDG uptake. In the SK-MEL-2 xenograft model, RO5126766 also suppresses the tumor growth. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-MEKR / MEK / p-ERK / ERK / p27 / ppRB / pRB / Cyclin D1 / Cyclin E |
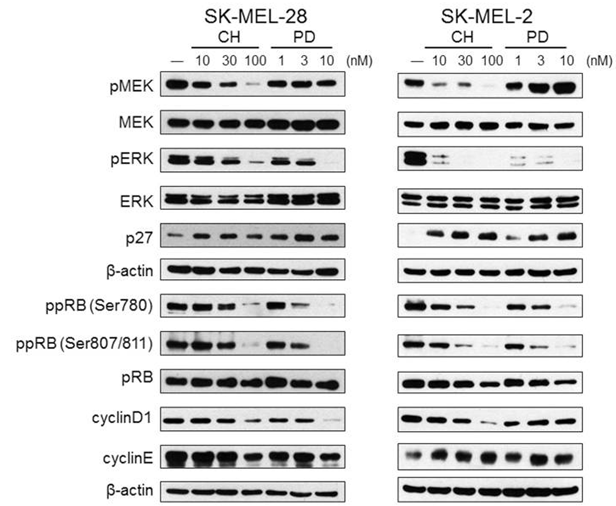
|
25422890 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT06104488 | Recruiting | Refractory Cancer|CNS Tumors|CNS Tumor Adult|CNS Tumor Childhood|MAP Kinase Family Gene Mutation|NF1|Plexiform Neurofibroma|Low-grade Glioma|Optic Pathway Gliomas|Neuroblastoma|Primary Brain Tumor|Solid Tumor|Solid Tumor Adult|Solid Carcinoma|Central Nervous System Tumor |
Memorial Sloan Kettering Cancer Center |
October 20 2023 | Phase 1 |
| NCT05375994 | Recruiting | Non Small Cell Lung Cancer|KRAS Activating Mutation|Advanced Cancer|Metastatic Cancer|Malignant Neoplasm of Lung|Malignant Neoplastic Disease |
Verastem Inc.|Mirati Therapeutics Inc. |
August 1 2022 | Phase 1|Phase 2 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































