
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
Infigratinib (BGJ398) FGFR inhibitor
Cat.No.S2183
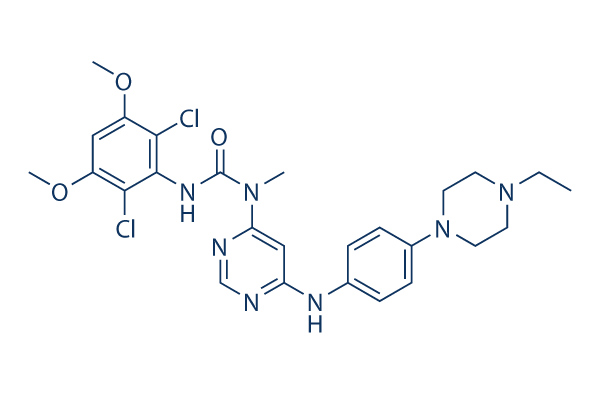
Chemical Structure
Molecular Weight: 560.48
Quality Control
Batch:
Purity:
99.87%
99.87
Products Often Used Together with Infigratinib (BGJ398)
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| HCC | Growth Inhibition Assay | 1-2500 nM | 48 h | IC50= 2359 nm | 25688743 | |
| HCC | Growth Inhibition Assay | 1-2500 nM | 48 h | IC50=1124 nm | 25688743 | |
| HCT116 | Growth Inhibition Assay | 48 h | IC50=3 μM | 24503538 | ||
| HKH2 | Growth Inhibition Assay | 48 h | IC50=4 μM | 24503538 | ||
| RKO | Growth Inhibition Assay | 48 h | IC50=1.2 μM | 24503538 | ||
| LS174T | Growth Inhibition Assay | 48 h | IC50=4 μM | 24503538 | ||
| HCD9 | Growth Inhibition Assay | 0.5-5 μM | 48/72 h | DMSO | decreases cell viability | 24135816 |
| HCT116 | Growth Inhibition Assay | 0.5-5 μM | 48/72 h | DMSO | decreases cell viability | 24135816 |
| SNU-C1 | Growth Inhibition Assay | 0.5-5 μM | 48/72 h | DMSO | no effect | 24135816 |
| MFE280 | Growth Inhibition Assay | IC50=2.63 ± 0.82 μM | 23443805 | |||
| AN3CA | Growth Inhibition Assay | IC50=1.00 ± 0.20 μM | 23443805 | |||
| HEC155 | Growth Inhibition Assay | IC50=4.74 ± 1.09 μM | 23443805 | |||
| MFE296 | Growth Inhibition Assay | IC50=2.86 ± 0.20 μM | 23443805 | |||
| SPAC1S | Growth Inhibition Assay | IC50=3.19 ± 0.93 μM | 23443805 | |||
| RL952 | Growth Inhibition Assay | IC50=3.41 ± 0.23 μM | 23443805 | |||
| EN1 | Growth Inhibition Assay | IC50=4.75 ± 0.62 μM | 23443805 | |||
| SNGII | Growth Inhibition Assay | IC50=4.29 ± 0.58 μM | 23443805 | |||
| ISHIKAWA | Growth Inhibition Assay | IC50=5.48 ± 0.03 μM | 23443805 | |||
| HEC1A | Growth Inhibition Assay | IC50=10.00 ± 1.00 μM | 23443805 | |||
| KLE | Growth Inhibition Assay | IC50=3.03 ± 0.11 μM | 23443805 | |||
| SNGM | Growth Inhibition Assay | IC50=5.00 ± 0.41 μM | 23443805 | |||
| USPC2 | Growth Inhibition Assay | IC50=7.00 ± 0.21 μM | 23443805 | |||
| EN | Growth Inhibition Assay | IC50=6.03 ± 0.31 μM | 23443805 | |||
| MFE319 | Growth Inhibition Assay | IC50=5.37 ± 0.03 μM | 23443805 | |||
| EFE184 | Growth Inhibition Assay | IC50=8.04 ± 0.69 μM | 23443805 | |||
| ECC1 | Growth Inhibition Assay | IC50=6.74 ± 0.59 μM | 23443805 | |||
| HEC1B | Growth Inhibition Assay | IC50=6.45 ± 0.67 μM | 23443805 | |||
| USPC1 | Growth Inhibition Assay | IC50=5.75 ± 0.50 μM | 23443805 | |||
| SPAC1L | Growth Inhibition Assay | IC50=4.92 ± 0.50 μM | 23443805 | |||
| HCC827 | Function assay | Displacement of [3H]-cyclopamine from SMO V404M mutant in gefitinib resistant human HCC827 cells by scintillation counting, Ki = 0.0465 μM. | 28787156 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| sf9 | Function assay | 60 mins | Inhibition of non-phosphorylated N-terminal His6-tagged FGFR4 C552A mutant (G442 to E753 residues) (unknown origin) expressed in sf9 cells using 5-Fluo-Ahx-KKKKEEIYFFFG-NH2 as substrate after 60 mins by microfluidic mobility shift assay, IC50 = 0.00082 μM. | ChEMBL | ||
| sf9 | Function assay | 60 mins | Inhibition of wild type non-phosphorylated N-terminal His6-tagged FGFR4 (G442 to E753 residues) (unknown origin) expressed in sf9 cells using 5-Fluo-Ahx-KKKKEEIYFFFG-NH2 as substrate after 60 mins by microfluidic mobility shift assay, IC50 = 0.062 μM. | ChEMBL | ||
| sf9 | Function assay | 60 mins | Inhibition of non-phosphorylated N-terminal His6-tagged FGFR4 C477A mutant (G442 to E753 residues) (unknown origin) expressed in sf9 cells using 5-Fluo-Ahx-KKKKEEIYFFFG-NH2 as substrate after 60 mins by microfluidic mobility shift assay, IC50 = 0.064 μM. | ChEMBL | ||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 560.48 | Formula | C26H31Cl2N7O3 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 872511-34-7 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | NVP-BGJ398 | Smiles | CCN1CCN(CC1)C2=CC=C(C=C2)NC3=CC(=NC=N3)N(C)C(=O)NC4=C(C(=CC(=C4Cl)OC)OC)Cl | ||
Solubility
|
In vitro |
DMSO
: 3 mg/mL
(5.35 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki | |
|---|---|
| In vitro |
Infigratinib (BGJ398) also prevents VEGFR2 with low potency, with an IC50 of 0.18 μM for inhibiting this target. It suppresses other kinases including ABL, FYN, KIT, LCK, LYN and YES with IC50 values of 2.3 μM, 1.9 μM, 0.75 μM, 2.5 μM, 0.3 μM and 1.1 μM, respectively. At the cellular level, this compound inhibits the proliferation of the FGFR1-, FGFR2-Q, and FGFR3-dependent BaF3 cells with IC50 of 2.9 μM, 2.0 μM and 2 μM, respectively. It interferes with autophosphorylation on specific tyrosine residues including FGFR-WT, FGFR2-WT, FGFR3-K650E, FGFR3-S249C and FGFR4-WT with IC50 of 4.6 nM, 4.9 nM, 5 nM, 5 nM and 168 nM, respectively. Additionally, it suppresses proliferation of the cancer cells with wild-type (WT) FGFR3 overexpression such as RT112, RT4, SW780 and JMSU1 with IC50 of 5 nM, 30 nM, 32 nM and 15 nM, respectively. [1]
|
| Kinase Assay |
Radiometric kinase assay
|
|
The enzymatic kinase activity is assessed by measuring the phosphorylation of a synthetic substrate by the purified GST-fusion FGFR3-K650E kinase domain, in the presence of radiolabeled ATP. Enzyme activities are measured by mixing 10 μL of a 3-fold concentrated solution of Infigratinib (BGJ398) or control with 10 μL of the corresponding substrate mixture (peptidic substrate, ATP and [γ33P]ATP). The reactions are initiated by addition of 10 μL of a 3-fold concentrated solution of the enzyme in assay buffer. The final concentrations of the assay components are as following: 10 ng of GST-FGFR3-K650E, 20 mM Tris-HCl, pH 7.5, 3 mM MnCl2, 3 mM MgCl2, 1 mM DTT, 250 μg/mL PEG 20000, 2 μg/mL poly(EY) 4:1, 1% DMSO and 0.5 μM ATP (γ-[33P]-ATP 0.1 μCi). The assay is carried out according to the filter binding (FB) method in 96-well plates at room temperature for 10 minutes in a final volume of 30 μL including this compound. The enzymatic reactions are stopped by the addition of 20 μL of 125 mM EDTA, and the incorporation of 33P into the polypeptidic substrates is quantified as following: 30 μL of the stopped reaction mixture are transferred onto Immobilon-PVDF membranes previously soaked for 5 minutes with methanol, rinsed with water, soaked for 5 min with 0.5% H3PO4, and mounted on vacuum manifold with disconnected vacuum source. After spotting, vacuum is connected, and each well rinsed with 0.5% H3PO4 (200 μL). Free membranes are removed and ished four times on a shaker with 1% H3PO4 and once with ethanol. Membranes are dried and overlaid with addition of 10 μL/well of a scintillation fluid. The plates are eventually sealed and counted in a microplate scintillation counter. IC50 values are calculated by linear regression analysis of the percentage inhibition of it.
|
|
| In vivo |
In this orthotopic xenograft bladder cancer model, BGJ398 induces tumor growth inhibition and stasis after oral administration for 12 consecutive days at the doses of 10 and 30 mg/kg, respectively. Interestingly, the animals that received this compound exhibit either no body weight loss (10 mg/kg) or 10% body weight gain (30 mg/kg), a further indication of efficacy. RT112 tumor-bearing and female Rowett rats receive a single oral administration of the monophosphate salt of Infigratinib (BGJ398) at the doses of 4.25 and 8.51 mg/kg. It significantly decreases the levels of pFRS2 and pMAPK in a dose-dependent manner. Additionally, it inhibits significantly bFGF-stimulated angiogenesis in a dose-dependent manner. However, it does not impair VEGF-induced blood vessel formation. [1]
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | pFGFR1 / FGFR1 / pFRS2 / FRS2 / MEK / ERK p-YAP (S127) / YAP Mcl-1 Cyclin D1 / Cyclin A / Cyclin B / γ-H2AX / Cleaved caspase-9 / PARP Snail / Slug / ZEB1 p-FRS2 / FRS2 / p-AKT / AKT / p-ERK / ERK |
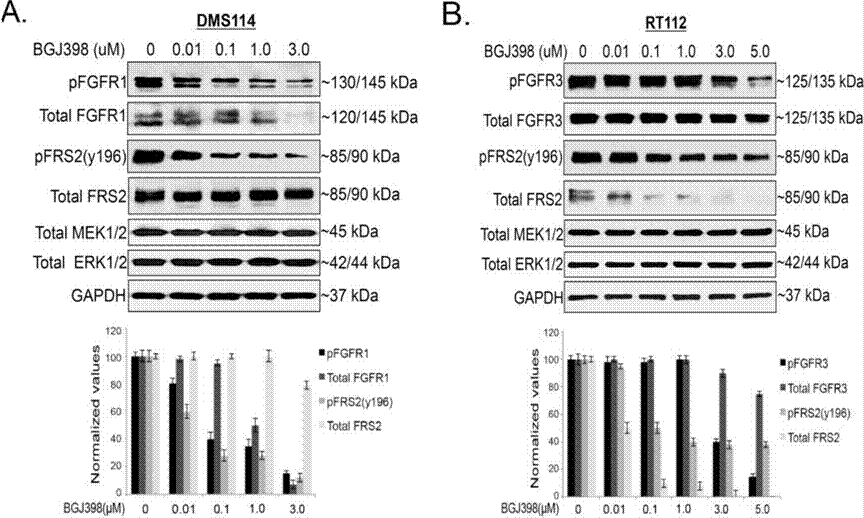
|
28255027 |
| Immunofluorescence | YAP |
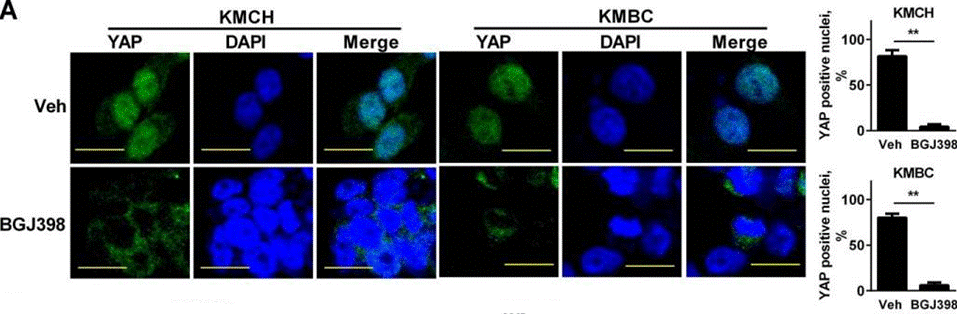
|
26826125 |
| Growth inhibition assay | Cell viability IC50 |
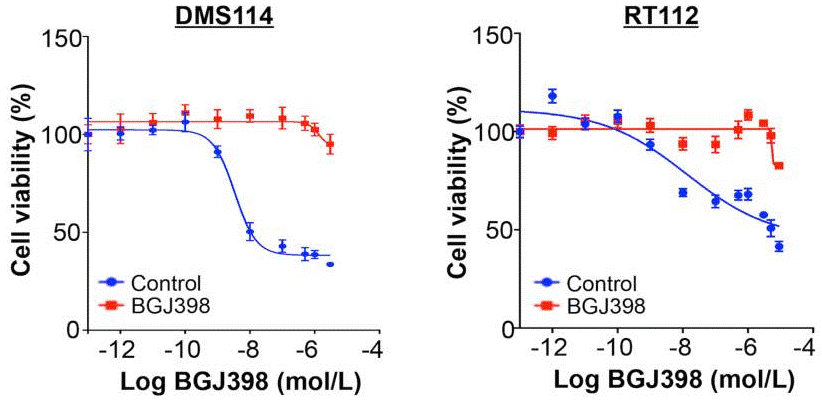
|
28255027 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT03510455 | Terminated | Tumor-Induced Osteomalacia|Oncogenic Osteomalacia |
National Institute of Dental and Craniofacial Research (NIDCR)|National Institutes of Health Clinical Center (CC) |
February 27 2019 | Phase 2 |
| NCT02312804 | Withdrawn | Cancer of Cervix|Tumors |
The University of Texas Health Science Center at San Antonio |
January 2015 | Phase 1 |
| NCT02160041 | Terminated | Solid Tumor|Hematologic Malignancies |
Novartis Pharmaceuticals|Novartis |
July 24 2014 | Phase 2 |
| NCT01928459 | Completed | Advanced Solid Tumors|Metastatic Solid Tumors |
Novartis Pharmaceuticals|Novartis |
October 2013 | Phase 1 |
| NCT01004224 | Completed | Advanced Solid Tumors With Alterations of FGFR1 2 and or 3|Squamous Lung Cancer With FGFR1 Amplification|Bladder Cancer With FGFR3 Mutation or Fusion|Advanced Solid Tumors With FGFR1 Amplication|Advanced Solid Tumors With FGFR2 Amplication|Advanced Solid Tumors With FGFR3 Mutation |
Novartis Pharmaceuticals|Novartis |
December 11 2009 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
If you have any suggestions about the formulation of this compound for a direct oral gavage administration?
Answer:
It can be dissolved in 30% PEG400/0.5% Tween80/5% Propylene glycol at 30 mg/ml as a suspension.






































