research use only
JIB-04 Histone Demethylase inhibitor
Cat.No.S7281
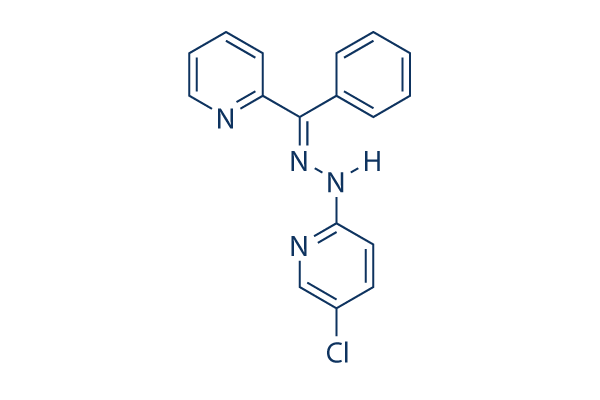
Chemical Structure
Molecular Weight: 308.76
Quality Control
| Related Targets | HDAC JAK BET Histone Methyltransferase PKC PARP HIF PRMT EZH2 AMPK |
|---|---|
| Other Histone Demethylase Inhibitors | GSK-J4 Hydrochloride SP2509 GSK-LSD1 2HCl Ladademstat (ORY-1001) Dihydrochloride CPI-455 HCl OG-L002 IOX1 GSK J1 ML324 CPI-455 |
Solubility
|
In vitro |
DMSO
: 62 mg/mL
(200.8 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 308.76 | Formula | C17H13ClN4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 199596-05-9 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | NSC 693627 | Smiles | C1=CC=C(C=C1)C(=NNC2=NC=C(C=C2)Cl)C3=CC=CC=N3 | ||
Mechanism of Action
| Targets/IC50/Ki |
JARID1A
(Cell-free assay) 230 nM
JMJD2D
(Cell-free assay) 290 nM
JMJD2E
(Cell-free assay) 340 nM
JMJD2B
(Cell-free assay) 435 nM
JMJD2A
(Cell-free assay) 445 nM
JMJD3
(Cell-free assay) 855 nM
|
|---|---|
| In vitro |
JIB-04 induces transcriptional changes in a cancer-selective manner, including the downregulation of proliferative genes and the upregulation of the anti-proliferative/pro-apoptotic genes. This compound blocks growth of lung and prostate cancer lines with IC50 as low as 10 nM, while produces less anti-proliferative activities on HBECs and PrSCs/PrECs.
|
| Kinase Assay |
Jumonji demethylase/prolyl hydroxylase/LSD1 activity assays
|
|
Active JMJD2E aa 1-350 is purified from E.coli and used in vitro in the presence of α-ketoglutarate, 5-10 μM iron, ascorbic acid and a histone peptide substrate in a coupled reaction with formaldehyde dehydrogenase supplemented with NAD+ to quantify NADH production or using Epigentek kit P-3081. For histone demethylation reactions quantified by Western analysis, a His-tagged hJMJ2D aa 1-350 expression construct, the kind gift of Drs. Y. Shi and J. Whetstine, is expressed and purified from E.coli following the Qiagen Ni-NTA agarose manual instructions and the protocol of Whestine et al 54. Briefly, protein is eluted in 50 mM TrisHCl pH 7.8 + 0.3 M NaCl + 10% Glycerol + 200 mM immidazol, and dialyzed against 20 mM TrisHCl pH 7.4, 0.15 M NaCl, 0.2 mM PMSF, 0.5 mM DTT, 8% glycerol. Enzyme is aliquoted, flash frozen and stored at -80°C. For activity assays by Western blot, ~1.5 μg of enzyme are combined with 0.3 μg H3K9me3 substrate (Active Motif #31213) in 10 μM (NH4)2Fe(SO4)2, 1 mM α-ketoglutarate, and 2 mM sodium L-ascorbate in 50 mM Hepes pH 7.9 in the presence of vehicle or drug and incubated for 30 min-2 hrs at 37°C. SDS loading buffer is added to the reactions, and after boiling, samples are run on NuPage 4-12% Bis-Tris gels, transferred to nitrocellulose and blotted using Upstate #07-523 to detect H3K9me3. For the detection of H3 total signal, we uses Active Motif #39763 (primary) and IRDye 680 conjugated donkey anti-rabbit IgG (secondary, LI-COR # 926-32223) and imaged blots in an Odyssey Infrared Imaging system kindly made available by Dr. M. Cobb. For in vitro IC50 determinations and competition studies, typically 100-200 ng of purified protein are incubated with vehicle, JIB-04 or analogs, as indicated in figure legends and activity measured by ELISA (Epigentek kit P-3081 for H3K9me3 demethylation, P-3083 for H3K4me3 demethylation, and P-3085 for H3K27me3 demethylation) in reactions containing 50mM Hepes pH 7.5, 0.01% Tween 20, 120nM (NH4)2Fe(SO4)2, 1 mM α-ketoglutarate, 2 mM sodium L-ascorbate and 50ng peptide substrate. Final enzyme concentrations in the reactions were as follows: 206 nM JMJD2A, 12 nM JMJD2B, 60 nM JMJD2C, 90 nM JMJD2D E.coli, 30 nM JMJD2D Sf9, 30 nM JMJD2E, 30 nM Jarid1a, 35 nM JMJD3. Background readings are given by heat inactivated enzymes, 0.5-1 mM 2,4 PDCA or reactions with no 2-OG. hJMJD2A (aa1-350) purified in E.coli is the kind gift of Dr. Jose Rizo-Rey and is assayed at 400 ng/reaction due to its intrinsic low activity. GraphPad Prism software is used for IC50 calculations and curve fitting. E.coli JMJD2D purified by us and Sf9 JMJD2D from BPS gave undistinguishable results. Note that for substrate competition assays, in order to remain in the linear range of the assay and not saturate binding capacity of the ELISA plate, reactions with > 0.75μM H3K9me3 containes unbiotinylated substrate or are diluted at the detection step and signals adjusted per dilution factor. For the direct quantification of H3K9me3 demethylase activity in cell lysates, treated cells (plated at 2 million/10cm dish) or tumor homogenates in PBS were sonicated (3x 4 sec) and equal amounts of protein are incubated with a histone H3K9me3 substrate in a reaction buffer containing cofactors for 2h at 37°C before specific immune-detection of the H3K9me2 product using Epigentek kit P-3081 reagents. 500 ng of E.coli purified PHD2 protein in 40mM Tris pH 7.4, 100mM NaCl, 20% glycerol, 5mM β-mercapto-ethanol, 10mM maltose are used to obtain activity in the linear range. Biotinylated peptides derived from the HIF-1 ODD (Biotin-Acp-DLDLEALAPYIPADDDFQL or Biotin-Acp-DLDLEALAP(OH)YIPADDDFQL as a hydroxylated control) are immobilized on Neutravidin-coated 96-well plates. Enzyme is incubated in the coated wells in reaction buffer (20 mM Tris-Cl pH 7.5, 5 mM KCl, 1.5 mM MgCl2, 2 mM DTT, 0.12 μM ferrous sulfate, 0.5 mM 2-oxoglutarate and 1 mM ascorbate) for 45 min at room temperature in the presence of the indicated drugs. The competitive analog of α-ketoglutarate, DMOG, is used as a positive control for inhibition. Peptide hydroxylation is detected using a polyclonal rabbit antibody raised against a hydroxylated HIF peptide epitope, (rabbit anti-hydroxyproline 4817, made in house), followed by addition of a goat anti-rabbit HRP-conjugated secondary antibody. Luminescence is measured in an EnVision plate reader. The activity of LSD1 recombinant protein is measured using Epigentek kit P-3075 according to the manufacturer’s protocol with the proprietary inhibitor.
|
|
| In vivo |
In two separate xenograft mouse models (H358 or A549), JIB-04 diminishes tumor growth, lowers Jumonji histone demethylase activity in tumors, and prolongs cancer survival.
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | H3K9me2 / H3K9me3 / H3K27me3 / H3K4me3 / TIMP1 / ANP αCD133 |
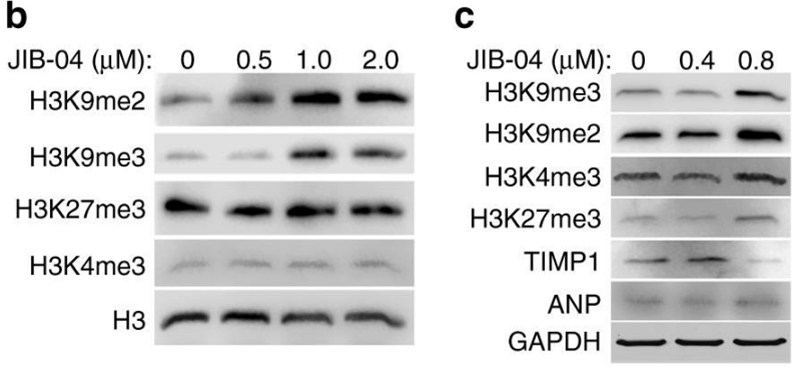
|
30531796 |
| Immunofluorescence | DNA-PKcs |
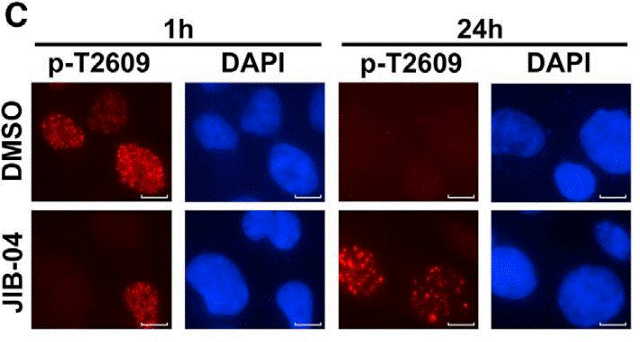
|
30355483 |
| Growth inhibition assay | Cell viability |
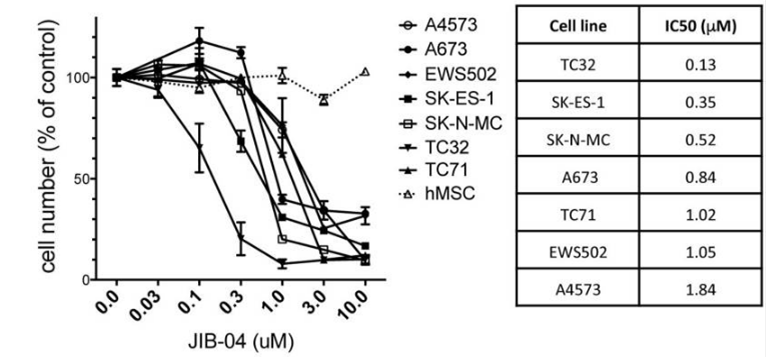
|
30237855 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
Do you have any suggestion for in vivo study please? (via injection)
Answer:
For IP or sub cutaneous injection, this compound is soluble in 4% DMSO+30% PEG 300+5% Tween 80+ddH2O at 3 mg/ml. But when water added, it turned yellowish green immediately. We are not sure if it was still fine in this vehicle. So we tried to use oil. 3mg of it dissolves in 40ul of DMSO clearly, and it can be dilute with corn oil at any proportion.






































