research use only
PHA-665752 c-Met inhibitor
Cat.No.S1070
Chemical Structure
Molecular Weight: 641.61
Quality Control
| Related Targets | EGFR VEGFR PDGFR FGFR Src MEK CSF-1R FLT3 HER2 c-Kit |
|---|---|
| Other c-Met Inhibitors | Tepotinib Dihexa SGX-523 Foretinib SU11274 BMS-777607 JNJ-38877605 Tivantinib PF-04217903 Savolitinib (AZD6094) |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| NCI-SNU-5 | Growth Inhibition Assay | IC50=0.12375 μM | ||||
| LB2241-RCC | Growth Inhibition Assay | IC50=0.15702 μM | ||||
| KINGS-1 | Growth Inhibition Assay | IC50=0.35911 μM | ||||
| ALL-PO | Growth Inhibition Assay | IC50=0.81277 μM | ||||
| SK-LMS-1 | Growth Inhibition Assay | IC50=0.89846 μM | ||||
| MV-4-11 | Growth Inhibition Assay | IC50=1.2947 μM | ||||
| SUP-T1 | Growth Inhibition Assay | IC50=2.13964 μM | ||||
| MRK-nu-1 | Growth Inhibition Assay | IC50=2.40056 μM | ||||
| ES1 | Growth Inhibition Assay | IC50=3.34866 μM | ||||
| NOS-1 | Growth Inhibition Assay | IC50=4.39867 μM | ||||
| KM12 | Growth Inhibition Assay | IC50=4.418 μM | ||||
| Becker | Growth Inhibition Assay | IC50=5.2466 μM | ||||
| NCI-SNU-1 | Growth Inhibition Assay | IC50=5.63733 μM | ||||
| EW-22 | Growth Inhibition Assay | IC50=7.73614 μM | ||||
| ES6 | Growth Inhibition Assay | IC50=7.8195 μM | ||||
| A498 | Growth Inhibition Assay | IC50=8.28446 μM | ||||
| EW-16 | Growth Inhibition Assay | IC50=9.6543 μM | ||||
| CTV-1 | Growth Inhibition Assay | IC50=9.85024 μM | ||||
| ETK-1 | Growth Inhibition Assay | IC50=10.2931 μM | ||||
| NCI-H1395 | Growth Inhibition Assay | IC50=10.8024 μM | ||||
| DOHH-2 | Growth Inhibition Assay | IC50=10.9264 μM | ||||
| GI-1 | Growth Inhibition Assay | IC50=11.8596 μM | ||||
| HT-144 | Growth Inhibition Assay | IC50=14.2163 μM | ||||
| ES5 | Growth Inhibition Assay | IC50=14.467 μM | ||||
| NALM-6 | Growth Inhibition Assay | IC50=15.2196 μM | ||||
| KNS-81-FD | Growth Inhibition Assay | IC50=15.5849 μM | ||||
| TE-15 | Growth Inhibition Assay | IC50=16.5771 μM | ||||
| SCC-15 | Growth Inhibition Assay | IC50=18.3498 μM | ||||
| EoL-1-cell | Growth Inhibition Assay | IC50=18.4545 μM | ||||
| NCI-H720 | Growth Inhibition Assay | IC50=18.771 μM | ||||
| NB14 | Growth Inhibition Assay | IC50=19.5425 μM | ||||
| KE-37 | Growth Inhibition Assay | IC50=19.8233 μM | ||||
| LXF-289 | Growth Inhibition Assay | IC50=19.8629 μM | ||||
| RPMI-8402 | Growth Inhibition Assay | IC50=20.3269 μM | ||||
| SK-N-DZ | Growth Inhibition Assay | IC50=21.2131 μM | ||||
| ACN | Growth Inhibition Assay | IC50=22.2497 μM | ||||
| TE-11 | Growth Inhibition Assay | IC50=26.069 μM | ||||
| COLO-800 | Growth Inhibition Assay | IC50=27.17 μM | ||||
| MOLT-13 | Growth Inhibition Assay | IC50=27.1847 μM | ||||
| 697 | Growth Inhibition Assay | IC50=28.7633 μM | ||||
| VA-ES-BJ | Growth Inhibition Assay | IC50=29.3729 μM | ||||
| EW-13 | Growth Inhibition Assay | IC50=29.5045 μM | ||||
| NB7 | Growth Inhibition Assay | IC50=32.2665 μM | ||||
| MONO-MAC-6 | Growth Inhibition Assay | IC50=32.8795 μM | ||||
| SW962 | Growth Inhibition Assay | IC50=33.4513 μM | ||||
| KS-1 | Growth Inhibition Assay | IC50=33.9481 μM | ||||
| KU812 | Growth Inhibition Assay | IC50=34.5702 μM | ||||
| IMR-5 | Growth Inhibition Assay | IC50=37.4318 μM | ||||
| BC-1 | Growth Inhibition Assay | IC50=38.033 μM | ||||
| NCI-H510A | Growth Inhibition Assay | IC50=38.2032 μM | ||||
| EW-18 | Growth Inhibition Assay | IC50=40.8303 μM | ||||
| CCRF-CEM | Growth Inhibition Assay | IC50=42.2797 μM | ||||
| HH | Growth Inhibition Assay | IC50=43.5069 μM | ||||
| NCI-H2171 | Growth Inhibition Assay | IC50=46.0272 μM | ||||
| LC-2-ad | Growth Inhibition Assay | IC50=49.1413 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 128 mg/mL
(199.49 mM)
Ethanol : 10 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 641.61 | Formula | C32H34Cl2N4O4S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 477575-56-7 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC1=C(NC(=C1C(=O)N2CCCC2CN3CCCC3)C)C=C4C5=C(C=CC(=C5)S(=O)(=O)CC6=C(C=CC=C6Cl)Cl)NC4=O | ||
Mechanism of Action
| Targets/IC50/Ki |
c-Met
(Cell-free assay) 9 nM
RON
(Cell-free assay) 68 nM
Flk1
(Cell-free assay) 200 nM
|
|---|---|
| In vitro |
PHA-665752 significantly inhibits c-Met kinase activity with Ki of 4 nM, and exhibits >50-fold selectivity for c-Met compared with various tyrosine and serine-threonine kinases. This compound potently inhibits the HGF-stimulated c-Met autophosphorylation with IC50 of 25-50 nM. It also significantly blocks HGF- and c-Met-dependent functions such as cell motility and cell proliferation with IC50 of 40-50 nM and 18-42 nM, respectively. In addition, this chemical potently inhibits HGF-stimulated or constitutive phosphorylation of mediators of downstream of c-Met such as Gab-1, ERK, Akt, STAT3, PLC-γ, and FAK in multiple tumor cell lines. It inhibits cell growth in TPR-MET-transformed BaF3 cells with IC50 of <60 nM, and inhibits constitutive cell motility and migration by 92.5% at 0.2 μM. Inhibition of c-Met by this compound (0.2 μM) also induces cell apoptosis of 33.1% and G1 cell cycle arrest with cells in G1 phase increasing from 42.4% to 77.0%. It can cooperate with rapamycin to inhibit cell growth of TPR-MET-transformed BaF3 cells and non-small cell lung cancer H441 cells.
|
| Kinase Assay |
In vitro enzyme assay
|
|
The c-Met kinase domain GST-fusion protein is used for the c-Met assay. The IC50 value of PHA-665752 for the inhibition of c-Met is based on phosphorylation of kinase peptide substrates or poly-glu-tyr in the presence of ATP and divalent cation (MgCl2 or MnCl2 10-20 mM). The linear range (i.e., the time period over which the rate remains equivalent to the initial rate) is determined for c-Met, and the kinetic measurement and IC50 determination are performed within this range.
|
|
| In vivo |
Administration of PHA-665752 induces a dose-dependent tumor growth inhibition of S114 xenografts by 20 %, 39% and 68%, at dose of 7.5, 15, and 30 mg/kg/day, respectively. This compound treatment significantly reduces the tumor growth of NCI-H69, NCI-H441 and A549 in mouse xenografts by 99%, 75%, and 59%, respectively. It also significantly inhibits angiogenesis by >85%, due to decreasing the production of vascular endothelial growth factor and increasing the production of the angiogenesis inhibitor thrombospondin-1.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | IDO p-Met / Met / p-Akt / Akt / p-ERK / ERK |
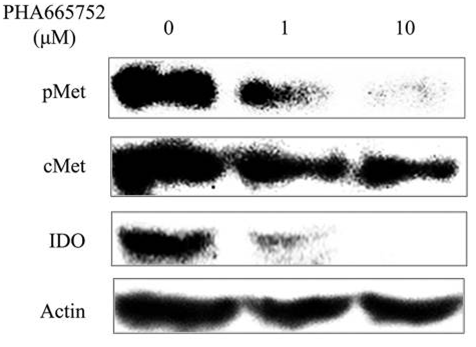
|
27082119 |
| Growth inhibition assay | Cell proliferation |
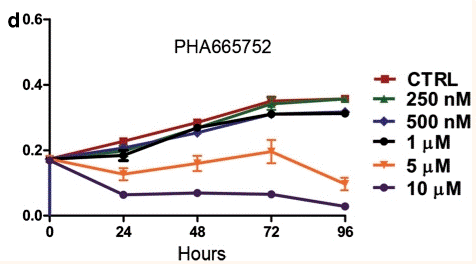
|
20603611 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
I need to use it for intraperitoneal application in mice. Could you tell me the solvent you use, please?
Answer:
The highest concentration of this compound (S1070) in 4% DMSO+30% PEG 300+5% Tween 80+ddH2O is 5mg/ml. If you want to get higher concentration, the concentration of DMSO and PEG will be higher. For example, it can be dissolved in 8% DMSO+50% PEG 300+5% Tween 80+ddH2O at 10mg/ml clearly.






































