research use only
PF-562271 Besylate FAK inhibitor
Cat.No.S2672
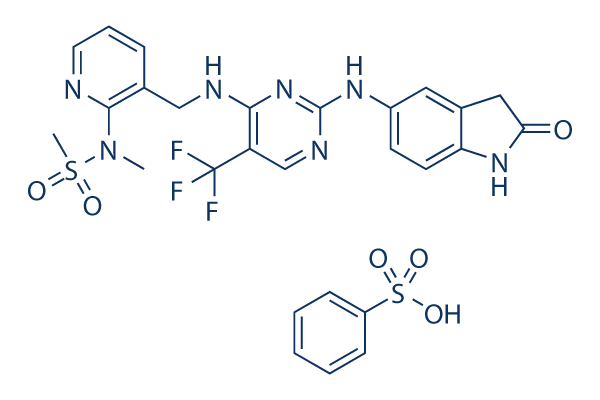
Chemical Structure
Molecular Weight: 665.66
Quality Control
| Related Targets | EGFR VEGFR JAK PDGFR FGFR Src HIF FLT FLT3 HER2 |
|---|---|
| Other FAK Inhibitors | Defactinib (VS-6063) PF-562271 (VS-6062) PF-573228 VS-4718 (PND-1186) PF-562271 HCl TAE226 (NVP-TAE226) GSK2256098 PF-431396 Y15 Ifebemtinib (IN10018, BI-853520) |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| MV-4-11 cell | Growth inhibition assay | Inhibition of human MV-4-11 cell growth in a cell viability assay, IC50=0.2766 μM | ||||
| human SW982 cell | Growth inhibition assay | Inhibition of human SW982 cell growth in a cell viability assay, IC50=0.3282 μM | ||||
| human KM12 cell | Growth inhibition assay | Inhibition of human KM12 cell growth in a cell viability assay, IC50=0.38557 μM | ||||
| human COLO-205 cell | Growth inhibition assay | Inhibition of human COLO-205 cell growth in a cell viability assay, IC50=0.48658 μM | ||||
| human COLO-829 cell | Growth inhibition assay | Inhibition of human COLO-829 cell growth in a cell viability assay, IC50=0.76176 μM | ||||
| human MG-63 cell | Growth inhibition assay | Inhibition of human MG-63 cell growth in a cell viability assay, IC50=0.80637 μM | ||||
| human IGROV-1 cell | Growth inhibition assay | Inhibition of human IGROV-1 cell growth in a cell viability assay, IC50=0.81038 μM | ||||
| human NCI-H650 cell | Growth inhibition assay | Inhibition of human NCI-H650 cell growth in a cell viability assay, IC50=0.83154 μM | ||||
| human RT-112 cell | Growth inhibition assay | Inhibition of human RT-112 cell growth in a cell viability assay, IC50=0.9846 μM | ||||
| human BCPAP cell | Growth inhibition assay | Inhibition of human BCPAP cell growth in a cell viability assay, IC50=1.01288 μM | ||||
| ALL-PO cell | Growth inhibition assay | Inhibition of human ALL-PO cell growth in a cell viability assay, IC50=1.01584 μM | ||||
| human KYSE-270 cell | Growth inhibition assay | Inhibition of human KYSE-270 cell growth in a cell viability assay, IC50=1.04714 μM | ||||
| human 8305C cell | Growth inhibition assay | Inhibition of human 8305C cell growth in a cell viability assay, IC50=1.09904 μM | ||||
| NCI-H810 cell | Growth inhibition assay | Inhibition of human NCI-H810 cell growth in a cell viability assay, IC50=1.10776 μM | ||||
| human CAL-33 cell | Growth inhibition assay | Inhibition of human CAL-33 cell growth in a cell viability assay, IC50=1.12938 μM | ||||
| human AN3-CA cell | Growth inhibition assay | Inhibition of human AN3-CA cell growth in a cell viability assay, IC50=1.21867 μM | ||||
| human NKM-1 cell | Growth inhibition assay | Inhibition of human NKM-1 cell growth in a cell viability assay, IC50=1.27506 μM | ||||
| human BPH-1 cell | Growth inhibition assay | Inhibition of human BPH-1 cell growth in a cell viability assay, IC50=1.28766 μM | ||||
| human MES-SA cell | Growth inhibition assay | Inhibition of human MES-SA cell growth in a cell viability assay, IC50=1.30682 μM | ||||
| human CAL-62 cell | Growth inhibition assay | Inhibition of human CAL-62 cell growth in a cell viability assay, IC50=1.31909 μM | ||||
| human KYSE-150 cell | Growth inhibition assay | Inhibition of human KYSE-150 cell growth in a cell viability assay, IC50=1.35236 μM | ||||
| human SK-UT-1 cell | Growth inhibition assay | Inhibition of human SK-UT-1 cell growth in a cell viability assay, IC50=1.44647 μM | ||||
| human HUTU-80 cell | Growth inhibition assay | Inhibition of human HUTU-80 cell growth in a cell viability assay, IC50=1.44886 μM | ||||
| human SIG-M5 cell | Growth inhibition assay | Inhibition of human SIG-M5 cell growth in a cell viability assay, IC50=1.48487 μM | ||||
| human AGS cell | Growth inhibition assay | Inhibition of human AGS cell growth in a cell viability assay, IC50=1.52124 μM | ||||
| human ST486 cell | Growth inhibition assay | Inhibition of human ST486 cell growth in a cell viability assay, IC50=1.53278 μM | ||||
| human HSC-2 cell | Growth inhibition assay | Inhibition of human HSC-2 cell growth in a cell viability assay, IC50=1.5395 μM | ||||
| human BC-1 cell | Growth inhibition assay | Inhibition of human BC-1 cell growth in a cell viability assay, IC50=1.61664 μM | ||||
| human CGTH-W-1 cell | Growth inhibition assay | Inhibition of human CGTH-W-1 cell growth in a cell viability assay, IC50=1.61679 μM | ||||
| human MZ1-PC cell | Growth inhibition assay | Inhibition of human MZ1-PC cell growth in a cell viability assay, IC50=1.62312 μM | ||||
| human SW1710 cell | Growth inhibition assay | Inhibition of human SW1710 cell growth in a cell viability assay, IC50=1.62628 μM | ||||
| human EW-13 cell | Growth inhibition assay | Inhibition of human EW-13 cell growth in a cell viability assay, IC50=1.63466 μM | ||||
| human U251 cell | Growth inhibition assay | Inhibition of human U251 cell growth in a cell viability assay, IC50=1.74031 μM | ||||
| human NCI-H460 cell | Growth inhibition assay | Inhibition of human NCI-H460 cell growth in a cell viability assay, IC50=2.04839 μM | ||||
| human DU-4475 cell | Growth inhibition assay | Inhibition of human DU-4475 cell growth in a cell viability assay, IC50=2.14759 μM | ||||
| human MFE-296 cell | Growth inhibition assay | Inhibition of human MFE-296 cell growth in a cell viability assay, IC50=2.47792 μM | ||||
| human DU-145 cell | Growth inhibition assay | Inhibition of human DU-145 cell growth in a cell viability assay, IC50=2.49118 μM | ||||
| human MDA-MB-231 cell | Growth inhibition assay | Inhibition of human MDA-MB-231 cell growth in a cell viability assay, IC50=2.49572 μM | ||||
| human SNU-387 cell | Growth inhibition assay | Inhibition of human SNU-387 cell growth in a cell viability assay, IC50=2.5282 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
4-Methylpyridine : 25 mg/mL
DMSO
: 0.4 mg/mL
(0.6 mM)
Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 665.66 | Formula | C21H20F3N7O3S.C6H6O3S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 939791-38-5 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | PF-00562271 Besylate | Smiles | CN(C1=C(C=CC=N1)CNC2=NC(=NC=C2C(F)(F)F)NC3=CC4=C(C=C3)NC(=O)C4)S(=O)(=O)C.C1=CC=C(C=C1)S(=O)(=O)O | ||
Mechanism of Action
| Targets/IC50/Ki |
FAK
(Cell-free assay) 1.5 nM
PYK2
(Cell-free assay) 13 nM
CDK2/CyclinE
(Cell-free assay) 30 nM
CDK3/CyclinE
(Cell-free assay) 47 nM
CDK1/CyclinB
(Cell-free assay) 58 nM
CDK7/CyclinH/MAT1
(Cell-free assay) 97 nM
FLT3
(Cell-free assay) 97 nM
GSK-3α
(Cell-free assay) 101 nM
CDK5/p35
(Cell-free assay) 120 nM
GSK-3β
(Cell-free assay) 133 nM
Aurora A
(Cell-free assay) 145 nM
|
|---|---|
| In vitro |
PF-562271 Besylate shows the selective inhibitory effects on FAK and Pyk2 tyrosine kinase activity with IC50 of 1.5 nM and 14 nM, respectively. And in cell-based assays, the IC50 of PF-562271 is shown to be 5 nM for FAK, which is more selective compared to other kinase targets. In 2 dimensional (2D) cultures, PF-562271 results in a dose-dependent cell proliferation inhibition in FAK WT, FAK−/− and FAK kinase-deficient (KD) cells with IC50 of 3.3 μM, 2.08 μM and 2.01 μM, respectively.
|
| Kinase Assay |
Recombinant kinase assay and enzyme kinetics
|
|
Briefly, purified-activated FAK kinase domain (amino acid 410–689) is reacted with 50 μM ATP and 10 μg per well of a random peptide polymer of Glu and Tyr, p(Glu/Tyr), in kinase buffer [50 mM HEPES (pH 7.5), 125 mM NaCl, and 48 mM MgCl2] for 15 minutes. Phosphorylation of p(Glu/Tyr) is challenged with serially diluted PF-562271 at 1/2-Log concentrations starting at a top concentration of 1 μM. Each concentration is tested in triplicate. Phosphorylation of p(Glu/Tyr) is detected with a general antiphospho-tyrosine (PY20) antibody followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody. HRP substrate is added, and absorbance readings at 450 nm are obtained after addition of stop solution (2 M H2SO4). IC50 values are determined using the Hill-Slope Model. Broad kinase selectivity profiling is performed in house and by using the KinaseProfiler Selectivity Screening Service available through UpState Biotechnology.
|
|
| In vivo |
In several human s.c. xenograft models, PF-562271 exhibits dose-dependent tumor growth inhibition, and produces maximum tumor inhibition for PC-3M, BT474, BxPc3, and LoVo ranging from 78% to 94% inhibition at doses of 25 to 50 mg/kg twice daily, without weight loss, morbidity, or death. PF-562271 (25 mg/kg by p.o.) leads to a significant decrease in tumor progression in both subcutaneous and bone metastasis PC3M-luc-C6 xenograft models. In a Huh7.5 hepatocellular carcinoma xenograft model, combination therapy of sunitinib and PF-562271 targets angiogenesis and tumor aggressiveness, and produces more significant anti-tumor effect than single agent by blocking tumor growth and impacting the ability of the tumor to recover upon withdrawal of the therapy.
|
References |
|
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT00666926 | Completed | Head and Neck Neoplasm|Prostatic Neoplasm|Pancreatic Neoplasm |
Verastem Inc. |
December 2005 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
We are planning both in vitro and in vivo experiments and want to know how to reconstitute it for these purposes?
Answer:
It has poor solubility in DMSO and water, with solubility in DMSO being only 0.4mg/ml. In a previous literature report (http://www.ncbi.nlm.nih.gov/pubmed/18339875), the author used 5% Gelucire to formulate this compound. You can also consider other co-solvents such as PEG400, CMC, Tween80, and Captisol.
Question 2:
Can you provide with a few common vehicles for it, S2672 for use as oral gavage?
Answer:
It can be dissolved in 0.5% CMC Na at 30 mg/ml as a suspension. If 4% DMSO can be used in your experiment, it will help dissolving the suspension more homogeneously.






































