research use only
GDC-0152 IAP antagonist
Cat.No.S7010
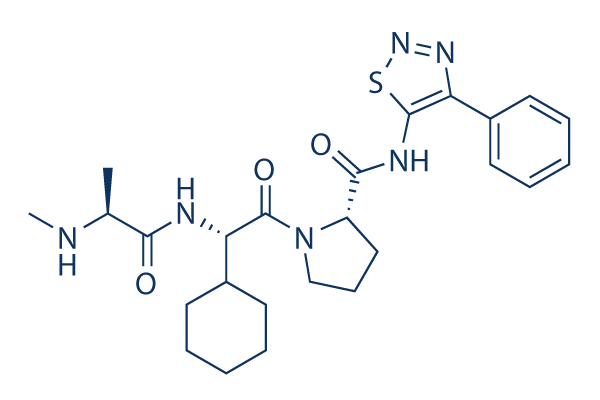
Chemical Structure
Molecular Weight: 498.64
Quality Control
| Related Targets | Bcl-2 Caspase PD-1/PD-L1 Ferroptosis p53 Apoptosis related Synthetic Lethality STAT TNF-alpha Ras |
|---|---|
| Other IAP Inhibitors | BV6 SM-164 Birinapant (TL32711) LCL161 Xevinapant (AT406) AZD5582 Tolinapant (ASTX660) |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| HEK293T cells | Function assay | 1-50 μM | 2 h | Inhibition of Flag-tagged XIAP BIR3 domain binding to cIAP1 expressed in human HEK293T cells at 1 to 50 uM after 2 hrs by immunoprecipitation | ||
| SK-MEL28 cells | Function assay | 0.5 μM | Inhibition of ML-IAP binding to Smax expressed in and zVAd treated human SK-MEL28 cells at 0.5 uM by immunoprecipitation | |||
| A2058 cells | Function assay | 15 mins | Induction of proteasomal degradation of cIAP1 in human A2058 cells after 15 mins by immunoblotting | |||
| MDA-MB-231 cells | Cytotoxicity assay | 72 h | Cytotoxicity against human MDA-MB-231 cells assessed as decrease in cell viability after 72 hrs by CellTiter-Glo luminescent assay | |||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 99 mg/mL
(198.54 mM)
Ethanol : 99 mg/mL Water : 3 mg/mL |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 498.64 | Formula | C25H34N6O3S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 873652-48-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC(C(=O)NC(C1CCCCC1)C(=O)N2CCCC2C(=O)NC3=C(N=NS3)C4=CC=CC=C4)NC | ||
Mechanism of Action
| Targets/IC50/Ki |
MLXBIR3SG
(Cell-free assay) 14 nM(Ki)
cIAP1-BIR3
(Cell-free assay) 17 nM(Ki)
XIAP-BIR3
(Cell-free assay) 28 nM(Ki)
cIAP2-BIR3
(Cell-free assay) 43 nM(Ki)
XIAP-BIR2
(Cell-free assay) 112 nM(Ki)
|
|---|---|
| In vitro |
GDC-0152 can block protein−protein interactions that involve IAP proteins and pro-apoptotic molecules. Using transiently transfected HEK293T cells, this compound is shown to disrupt XIAP binding to partially processed caspase-9 and to disrupt the association of ML-IAP, cIAP1, and cIAP2 with Smac. In melanoma SK-MEL28 cells, the endogenous association of ML-IAP and Smac is effectively also abolished by this chemical. It leads to a decrease in cell viability in the MDA-MB-231 breast cancer cell line, while having no effect on normal human mammary epithelial cells (HMEC). This compound is found to activate caspases 3 and 7 in a dose- and time-dependent manner. It is shown to induce rapid degradation of cIAP1 in A2058 melanoma cells. It effectively induces degradation of cIAP1 at concentrations as low as 10 nM, consistent with its affinity for cIAP1.
|
| Kinase Assay |
Fluorescence polarization-based competition assay
|
|
Inhibition constants ( Ki ) for the antagonists are determined by addition of the IAP protein constructs to wells containing serial dilutions of the antagonists or the peptide AVPW, and the Hid-FAM probe or AVP-diPhe-FAM probe, as appropriate, in the polarization buffer. Samples are read after a 30-minute incubation. Fluorescence polarization values are plotted as a function of the antagonist concentration, and the IC50 values are obtained by fitting the data to a 4-parameter equation using software. Ki values for the antagonists are determined from the IC50 valued.
|
|
| In vivo |
GDC-0152 has moderate predicted hepatic clearance based on metabolic stability assays conducted using human liver microsomes. Plasma−protein binding of this compound is moderate and comparable among mice (88−91%), rats (89−91%), dogs (81−90%), monkeys (76−85%), and humans (75−83%) over the range of concentrations investigated (0.1−100 μM); higher plasma−protein binding is observed in rabbits (95−96%). It does not preferentially distribute to red blood cells with blood−plasma partition ratios ranging from 0.6 to 1.1 in all species tested. The pharmacokinetics for this chemical is achieved with a C max of 53.7 μM and AUC of 203.5 h•μM.
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | cIAP1 / cIAP2 / XIAP / ML-IAP |
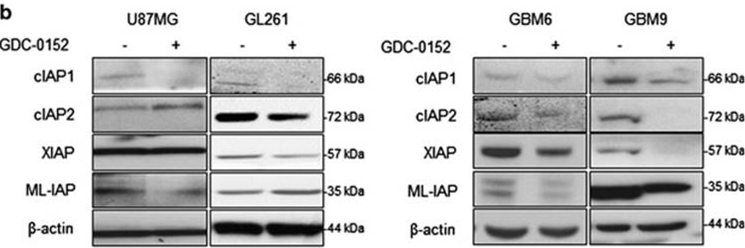
|
27490930 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT00977067 | Terminated | Solid Cancers |
Genentech Inc. |
June 2007 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































