research use only
CP-724714 HER2 inhibitor
Cat.No.S1167
Chemical Structure
Molecular Weight: 469.53
Quality Control
| Related Targets | EGFR VEGFR PDGFR FGFR c-Met Src MEK CSF-1R FLT3 c-Kit |
|---|---|
| Other HER2 Inhibitors | Sapitinib (AZD8931) Mubritinib (TAK 165) AC480 (BMS-599626) Tyrphostin AG 879 HER2-Inhibitor-1 Zongertinib TAS0728 |
Solubility
|
In vitro |
DMSO
: 94 mg/mL
(200.2 mM)
Ethanol : 94 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 469.53 | Formula | C27H27N5O3 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 537705-08-1 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CCOC(=O)NCC=CC1=CC2=C(C=C1)N=CN=C2NC3=CC(=C(C=C3)OC4=CN=C(C=C4)C)C | ||
Mechanism of Action
| Targets/IC50/Ki |
HER2/ErbB2
(Cell-free assay) 10 nM
|
|---|---|
| In vitro |
CP-724,714 is marked selectively against EGFR with IC50 of 6.4 μM. CP-724,714 is >1,000-fold less potent for IR, IGF-1R, PDGFRβ, VGFR2, abl. Src, c-Met c-jun NH2-terminal kinase (JNK)-2, JNK-3, ZAP-70, cyclin-dependent kinase (CDK)-2, and CDK-5. CP-724,714 potently reduces the EGF-induced autophosphorylation of the chimera containing the erbB2 kinase domain with IC50 of 32 nM, but is markedly less potent against EGFR in transfected NIH3T3 cells. CP-724,714 sensitively inhibits the proliferation of erbB2-amplified cells including BT-474 and SKBR3, with IC50 of 0.25 and 0.95 μM. CP-724,714 induces the accumulation of cells in G1 phase and a marked reduction in S-phase in BT-474 cells at 1 μM. CP-724,714 likely exerts its hepatotoxicity via both hepatocellular injury and hepatobiliary cholestatic mechanisms. CP-724,714 displays inhibition of cholyl-lysyl and taurocholate (TC) efflux into canaliculi in cryopreserved and fresh cultured human hepatocytes, respectively. CP-724,714 inhibits TC transport in membrane vesicles expressing human bile salt export pump with IC50 of 16 μM and inhibits the major efflux transporter in bile canaliculi, MDR1, with IC50 of ~28 μM. |
| Kinase Assay |
Kinase assays
|
|
Recombinant erbB2 (amino acid residues 675-1255) and EGFR (amino acid residues 668-1211) intracellular domains are expressed in baculovirus-infected Sf9 cells as S-transferase fusion proteins. The proteins are purified by affinity chromatography on Sepharose beads for use in the assay. Nunc MaxiSorp 96-well plates are coated by incubation overnight at 37 °C with 100 μL/well of 0.25 mg/mL poly(Glu:Tyr, 4:1), PGT in PBS. Excess PGT is removed by aspiration and the plate is washed 3 times with wash buffer (0.1% Tween 20 in PBS). The kinase reaction is performed in 50 μL of 50 mm HEPES (pH 7.4) containing 125 mm sodium chloride, 0.1 mm sodium orthovanadate, 1 mm ATP, and ∼15 ng of recombinant protein. Inhibitors in DMSO are added; the final DMSO concentration is 2.5%. Phosphorylation is initiated by addition of ATP and proceeded for 6 min at room temperature, with constant shaking. The kinase reaction is terminated by aspiration of the reaction mixture and washing four times with wash buffer. Phosphorylated PGT is measured after a 25-min incubation with 50 μL/well HRP conjugated-PY54 antiphosphotyrosine antibody, diluted to 0.2 μg/mL in blocking buffer (3% BSA, 0.05% Tween 20 in PBS). Antibody is removed by aspiration and the plate is washed four times with wash buffer. The colorimetric signal is developed by addition of 50 μL/well Tetramethylbenzidine Microwell Peroxidase Substrate and stopped by the addition of 50 μL/well 0.09 m sulfuric acid. The phosphotyrosine product formed is estimated by measurement of absorbance at 450 nm. The signal for controls is typically A0.6–1.2, with essentially no background in wells without ATP, kinase protein, or PGT, and is proportional to the time of incubation for 6 min.
|
|
| In vivo |
CP-724,714 (25 mg/kg) is rapidly absorbed after p.o. administration and causes reduction of tumor erbB2 receptor phosphorylation after dosing in FRE-erbB2 or BT-474 xenografts. CP-724,714 induces apoptosis in FRE-erbB2 xenograft–bearing (s.c.) mice and shows 50% tumor growth inhibition at 50 mg/kg, without weight loss or mortality. CP-724,714 also has great antitumor activity in MDA-MB-453, MDA-MB-231, LoVo (colon), and Colo-205 (colon) xenografts. Furthermore, CP-724,714 (30 or 100 mg/kg) reduces the extracellular signal–regulated kinase and Akt phosphorylation in BT-474 xenografts. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-HER2 / HER2 |
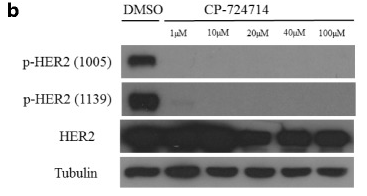
|
29490608 |
| Growth inhibition assay | Cell viability |
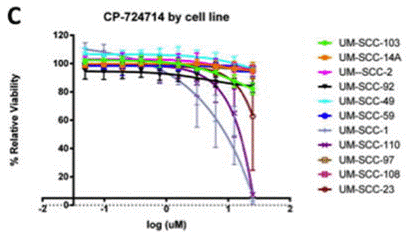
|
27077364 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































