research use only
Nodinitib-1 (ML130) NOD inhibitor
Cat.No.S2863
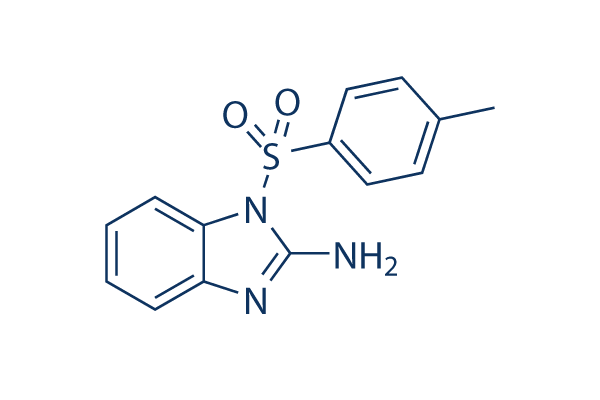
Chemical Structure
Molecular Weight: 287.34
Quality Control
| Related Targets | NF-κB HDAC Antioxidant ROS IκB/IKK Nrf2 AP-1 MALT |
|---|---|
| Other NOD Inhibitors | NOD-IN-1 Inarigivir soproxil |
Solubility
|
In vitro |
DMSO
: 57 mg/mL
(198.37 mM)
Ethanol : 2 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 287.34 | Formula | C14H13N3O2S |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 799264-47-4 | Download SDF | Storage of Stock Solutions |
|
|
Mechanism of Action
| Targets/IC50/Ki |
NOD1
0.56 μM
|
|---|---|
| In vitro |
Nodinitib-1 (ML130) has shown selective inhibition of NOD1-induced NF-κB activation in HEK293 cells with no cytotoxicity and is selected as a probe candidate molecule. It is also confirmed in secondary assays by selectively inhibiting NOD1-dependent IL-8 secretion and the NOD1-dependent pathway to NF-κB activation. In another research, this compound is proved to cause conformational changes of NOD1 in vitro and alter NOD1 subcellular targeting in cells, providing chemical probes for interrogating mechanisms regulating NOD1 activity and tools for exploring the roles of NOD1 in various infectious and inflammatory diseases. |
| Kinase Assay |
NOD1 Dose Response assay
|
|
Nodinitib-1 (ML130) 的测试流程如下:Day 1 Procedure 1) Harvest HEK-293-T NFKB-Luc at 100% confluency at 100% confluency. 2) Add NOD assay media with Multidrop. 3) Spin down plates at 1000 rpm for 1 min in centrifuge. 4) Serial compound dilutions. 5) Add gamma-tri-DAP to cell suspension at 0.75 ug/mL. 6) Seed 13000 cells/well in 4 uL/well to full plate HEK-293-T NFKB-Luc to TC-treated plate. 7) Spin down plates 500 RPM for 5 min on centrifuge. 8) Lid Plates. Sandwich 4 plates between 2 lidded 384 plates filled with H2O. 9) Wrap plates securely in single layer of Plastic Wrap. 10) Incubate overnight (14 hours) in 37 ℃ and 5% CO2 incubator. Day 2 Procedure 1) Add 3 ul/well of SteadyGlo solution with Multidrop. 2) Shake plates on a plate shaker for 20 min. 3) Spin plates 1000 RPM for 1 min using centrifuge. 4) Read luminescence.IC50 values are calculated using GraphPad Prism 5.0.The average Z
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-NF-κB / NF-κB / TNFα |
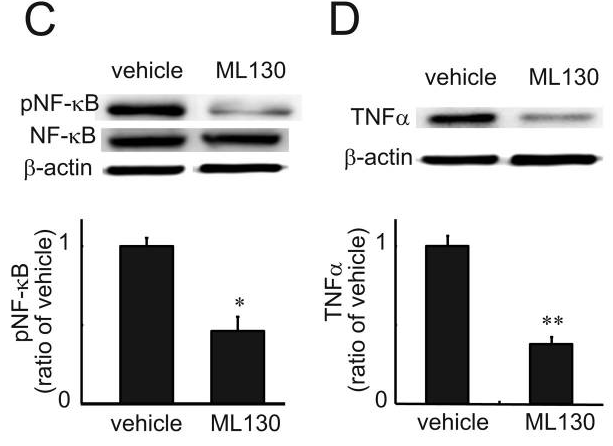
|
22450316 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































