research use only
ZM 447439 Aurora A/B Inhibitor
Cat.No.S1103
Chemical Structure
Molecular Weight: 513.59
Quality Control
| Related Targets | CDK HSP PD-1/PD-L1 ROCK Wee1 DNA/RNA Synthesis Microtubule Associated Ras KRas Casein Kinase |
|---|---|
| Other Aurora Kinase Inhibitors | Hesperadin Barasertib-HQPA (AZD2811) Alisertib (MLN8237) Tozasertib (VX-680) MLN8054 Danusertib (PHA-739358) MK-5108 TCS7010 (Aurora A Inhibitor I) AMG-900 PHA-680632 |
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human EoL-1-cell | Growth inhibition assay | Inhibition of human EoL-1-cell cell growth in a cell viability assay, IC50=0.18678 μM | ||||
| MCF7 cell | Proliferation assay | Antiproliferative activity against MCF7 cell line, IC50=0.198 μM | ||||
| human P12-ICHIKAWA cell | Growth inhibition assay | Inhibition of human P12-ICHIKAWA cell growth in a cell viability assay, IC50=0.22441 μM | ||||
| human KARPAS-45 cell | Growth inhibition assay | Inhibition of human KARPAS-45 cell growth in a cell viability assay, IC50=0.38128 μM | ||||
| human ES3 cell | Growth inhibition assay | Inhibition of human ES3 cell growth in a cell viability assay, IC50=0.4732 μM | ||||
| human ES8 cell | Growth inhibition assay | Inhibition of human ES8 cell growth in a cell viability assay, IC50=0.49806 μM | ||||
| human TE-11 cell | Growth inhibition assay | Inhibition of human TE-11 cell growth in a cell viability assay, IC50=0.53703 μM | ||||
| human RS4-11 cell | Growth inhibition assay | Inhibition of human RS4-11 cell growth in a cell viability assay, IC50=0.5544 μM | ||||
| human MOLT-16 cell | Growth inhibition assay | Inhibition of human MOLT-16 cell growth in a cell viability assay, IC50=0.60961 μM | ||||
| human RKO cell | Growth inhibition assay | Inhibition of human RKO cell growth in a cell viability assay, IC50=0.70656 μM | ||||
| human MV-4-11 cell | Growth inhibition assay | Inhibition of human MV-4-11 cell growth in a cell viability assay, IC50=0.7963 μM | ||||
| human SW954 cell | Growth inhibition assay | Inhibition of human SW954 cell growth in a cell viability assay, IC50=0.83635 μM | ||||
| human BE-13 cell | Growth inhibition assay | Inhibition of human BE-13 cell growth in a cell viability assay, IC50=0.84418 μM | ||||
| human MOLT-4 cell | Growth inhibition assay | Inhibition of human MOLT-4 cell growth in a cell viability assay, IC50=0.89978 μM | ||||
| human NBsusSR cell | Growth inhibition assay | Inhibition of human NBsusSR cell growth in a cell viability assay, IC50=0.91387 μM | ||||
| human H9 cell | Growth inhibition assay | Inhibition of human H9 cell growth in a cell viability assay, IC50=0.92979 μM | ||||
| human A172 cell | Growth inhibition assay | Inhibition of human A172 cell growth in a cell viability assay, IC50=0.98411 μM | ||||
| human ES5 cell | Growth inhibition assay | Inhibition of human ES5 cell growth in a cell viability assay, IC50=1.00248 μM | ||||
| human SBC-1 cell | Growth inhibition assay | Inhibition of human SBC-1 cell growth in a cell viability assay, IC50=1.03928 μM | ||||
| human NCI-H209 cell | Growth inhibition assay | Inhibition of human NCI-H209 cell growth in a cell viability assay, IC50=1.16602 μM | ||||
| human NKM-1 cell | Growth inhibition assay | Inhibition of human NKM-1 cell growth in a cell viability assay, IC50=1.16798 μM | ||||
| human NCI-H720 cell | Growth inhibition assay | Inhibition of human NCI-H720 cell growth in a cell viability assay, IC50=1.20627 μM | ||||
| human KE-37 cell | Growth inhibition assay | Inhibition of human KE-37 cell growth in a cell viability assay, IC50=1.21388 μM | ||||
| human SW48 cell | Growth inhibition assay | Inhibition of human SW48 cell growth in a cell viability assay, IC50=1.23155 μM | ||||
| human IST-SL1 cell | Growth inhibition assay | Inhibition of human IST-SL1 cell growth in a cell viability assay, IC50=1.31727 μM | ||||
| human SK-NEP-1 cell | Growth inhibition assay | Inhibition of human SK-NEP-1 cell growth in a cell viability assay, IC50=1.36498 μM | ||||
| human NOMO-1 cell | Growth inhibition assay | Inhibition of human NOMO-1 cell growth in a cell viability assay, IC50=1.36725 μM | ||||
| human DOHH-2 cell | Growth inhibition assay | Inhibition of human DOHH-2 cell growth in a cell viability assay, IC50=1.40276 μM | ||||
| human ABC-1 cell | Growth inhibition assay | Inhibition of human ABC-1 cell growth in a cell viability assay, IC50=1.40352 μM | ||||
| human Ramos-2G6-4C10 cell | Growth inhibition assay | Inhibition of human Ramos-2G6-4C10 cell growth in a cell viability assay, IC50=1.40605 μM | ||||
| human EM-2 cell | Growth inhibition assay | Inhibition of human EM-2 cell growth in a cell viability assay, IC50=1.41721 μM | ||||
| human NB14 cell | Growth inhibition assay | Inhibition of human NB14 cell growth in a cell viability assay, IC50=1.55621 μM | ||||
| human MOLT-13 cell | Growth inhibition assay | Inhibition of human MOLT-13 cell growth in a cell viability assay, IC50=1.56706 μM | ||||
| human ECC10 cell | Growth inhibition assay | Inhibition of human ECC10 cell growth in a cell viability assay, IC50=1.63353 μM | ||||
| human LK-2 cell | Growth inhibition assay | Inhibition of human LK-2 cell growth in a cell viability assay, IC50=1.64594 μM | ||||
| human CTB-1 cell | Growth inhibition assay | Inhibition of human CTB-1 cell growth in a cell viability assay, IC50=1.67082 μM | ||||
| human NCI-H1581 cell | Growth inhibition assay | Inhibition of human NCI-H1581 cell growth in a cell viability assay, IC50=1.6755 μM | ||||
| human COLO-800 cell | Growth inhibition assay | Inhibition of human COLO-800 cell growth in a cell viability assay, IC50=1.70382 μM | ||||
| human NB7 cell | Growth inhibition assay | Inhibition of human NB7 cell growth in a cell viability assay, IC50=1.75297 μM | ||||
| human LAMA-84 cell | Growth inhibition assay | Inhibition of human LAMA-84 cell growth in a cell viability assay, IC50=1.7552 μM | ||||
| human HCT-116 cells | Growth inhibition assay | Inhibition of human HCT-116 cell growth in a cell viability assay, IC50=1.80908 μM | ||||
| SK-UT-1 cell | Growth inhibition assay | Inhibition of human SK-UT-1 cell growth in a cell viability assay, IC50=1.8153 μM | ||||
| human H4 cell | Growth inhibition assay | Inhibition of human H4 cell growth in a cell viability assay, IC50=1.81578 μM | ||||
| human CAL-51 cell | Growth inhibition assay | Inhibition of human CAL-51 cell growth in a cell viability assay, IC50=1.83845 μM | ||||
| human LoVo cells | Proliferation assay | 72 h | Antiproliferative activity against human LoVo cells after 72 hrs by MTT-based WST8 reagent assay, IC50=1.9 μM | |||
| human HN cell | Growth inhibition assay | Inhibition of human HN cell growth in a cell viability assay, IC50=1.9251 μM | ||||
| human L-363 cell | Growth inhibition assay | Inhibition of human L-363 cell growth in a cell viability assay, IC50=1.9512 μM | ||||
| human NCI-H747 cell | Growth inhibition assay | Inhibition of human NCI-H747 cell growth in a cell viability assay, IC50=2.03353 μM | ||||
| human A498 cell | Growth inhibition assay | Inhibition of human A498 cell growth in a cell viability assay, IC50=2.3669 μM | ||||
| Click to View More Cell Line Experimental Data | ||||||
Solubility
|
In vitro |
DMSO
: 103 mg/mL
(200.54 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 513.59 | Formula | C29H31N5O4 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 331771-20-1 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | COC1=C(C=C2C(=C1)C(=NC=N2)NC3=CC=C(C=C3)NC(=O)C4=CC=CC=C4)OCCCN5CCOCC5 | ||
Mechanism of Action
| Features |
An Aurora selective ATP-competitive inhibitor.
|
|---|---|
| Targets/IC50/Ki |
Aurora A
(Cell-free assay) 110 nM
Aurora B
(Cell-free assay) 130 nM
LCK
(Cell-free assay) 880 nM
Src
(Cell-free assay) 1.03 μM
MEK1
(Cell-free assay) 1.79 μM
|
| In vitro |
In vitro, ZM-447439 selectively inhibits recombinant human Aurora A and B with IC50 values of 110 and 130 nM, respectively, while other protein kinases of diverse structural types including the mitotic kinases CDK1 and PLK1 are inhibited with IC50 values >10 μM. Aurora kinase inhibitor, ZM-447439 time- and dose-dependently inhibits the growth of all three cell lines with IC50 values of 3 μM (BON), 0.9 μM (QGP-1) and 3 μM (MIP-101) after 72 hours of continuous exposure. In addition, ZM-447439 potently induces cell apoptosis by promoting DNA fragmentation and caspase 3 and 7 activation, and arrests GEP-NET cells in the G0 /G1and G2/M phase of the cell cycle. In mouse embryo, inhibition of Aurora kinase activity by ZM-447439 results in abnormalities during mitosis by regulating the phosphorylation of histone H3 serine 10 (H3S10Ph) from G2 to metaphase with different perturbations in each embryonic cycle. A recent study shows that ZM-447439 exhibits growth inhibitory and proapoptotic effect on cervical cancer SiHa cells, and enhances the chemosensitivity. |
| Kinase Assay |
In vitro kinase assays
|
|
Recombinant Aurora A and B are expressed as NH2-terminal His6-tagged fusion proteins using a baculovirus expression system. Aurora A is purified by affinity chromatography using Ni-NTA agarose, and Aurora B is purified by ion exchange chromatography using CM Sepharose Fast Flow. 1 ng purified recombinant enzyme is added to a reaction cocktail containing 25 mM Tris-HCl, pH 7.5, 12.5 mM KCl, 2.5 mM NaF, 0.6 mM DTT, 6.25 mM MnCl2, 10 μM peptide substrate, 10 μM for Aurora A or 5 μM ATP for Aurora B, and 0.2 μCi γ-[33P]ATP (specific activity ≥2,500 Ci/mmol), and is then incubated at RT for 60 minutes. Reactions are stopped by addition of 20% phosphoric acid, and the products are captured on P30 nitrocellulose filters and assayed for incorporation of 33P with a BetaplateTM counter. No enzyme and no compound control values are used to determine the concentration of ZM447439, which gave 50% inhibition of enzyme activity. Further details are available on request from Nicholas Keen.
|
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p53 / p21 / p-p38 / p38 pHistone H3 / CEM BubR1 / MAD2 / Cyclin B1 |

|
23759594 |
| Immunofluorescence | p21 p53 Aurora B / Survivin |
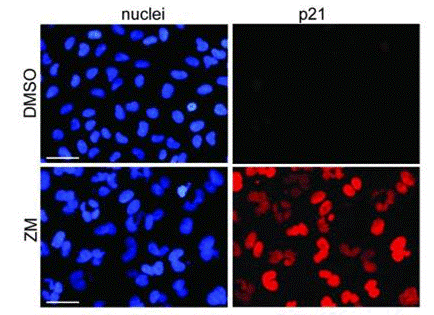
|
23759594 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































