- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- DNA Damage/DNA Repair
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
NSC 74859 (S3I-201)
NSC 74859 (S3I-201) shows potent inhibition of STAT3 DNA-binding activity with IC50 of 86 μM in cell-free assays, and low activity towards STAT1 and STAT5.
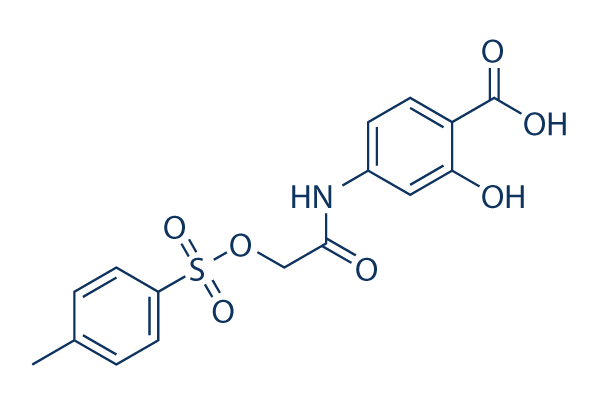
NSC 74859 (S3I-201) Chemical Structure
CAS No. 501919-59-1
Purity & Quality Control
Batch:
Purity:
99.28%
99.28
NSC 74859 (S3I-201) Related Products
| Related Targets | STAT1 STAT3 STAT5 STAT6 | Click to Expand |
|---|---|---|
| Related Products | Stattic Cryptotanshinone (Tanshinone C) SH-4-54 C188-9 BP-1-102 Napabucasin (BBI608) HO-3867 Nifuroxazide AS1517499 Homoharringtonine (HHT) STAT5-IN-1 Ochromycinone (STA-21) HJC0152 APTSTAT3-9R STAT3-IN-1 SC-1 SH5-07 (SH-5-07) Scutellarin Narciclasine | Click to Expand |
| Related Compound Libraries | Kinase Inhibitor Library FDA-approved Drug Library Natural Product Library Tyrosine Kinase Inhibitor Library JAK/STAT compound library | Click to Expand |
Signaling Pathway
Cell Data
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| U87 | Growth Inhibition Assay | 72 h | IC50=55.1 μM | 20072652 | ||
| U373 | Growth Inhibition Assay | 72 h | IC50=52.5 μM | 20072652 | ||
| HPAC | Growth Inhibition Assay | 72 h | IC50>100 μM | 20072652 | ||
| PANC-1 | Growth Inhibition Assay | 72 h | IC50>100 μM | 20072652 | ||
| SK-BR-3 | Growth Inhibition Assay | 72 h | IC50>100 μM | 20072652 | ||
| U-373 MG | Cytotoxicity Assay | 3/10 μM | 24 h | reduces FN-γ-induced cell neurotoxicity | 20888416 | |
| MDA-MB-231 | Growth Inhibition Assay | 72 h | IC50>100 μM | 20072652 | ||
| HUVEC | Function Assay | 0.5-20 μM | 24 h | DMSO | suppresses the hypoxia-induced accumulation of HIF-1α | 21523559 |
| Huh7 | Growth Inhibition Assay | 100 nM | 48 h | DMSO | inhibits the IL-6 stimulation promoted cell proliferation | 23364389 |
| PLC/PRF/5 | Growth Inhibition Assay | 100 nM | 48 h | DMSO | inhibits the IL-6 stimulation promoted cell proliferation | 23364389 |
| H460 | Function Assay | 50/100 μM | 48 h | inhibits the Stat3C increased miR-92a expression | 23820254 | |
| H1299 | Function Assay | 50/100 μM | 48 h | suppresses miR-92a expression dose-dependently | 23820254 | |
| T-cell | Growth Inhibition Assay | IC50=50 μM | 24068731 | |||
| U373 | Growth Inhibition Assay | 125 μM | 24 h | DMSO | disrupts STAT3 signaling and proliferation | 24070820 |
| HUT-102 | Apoptosis Assay | 75-300 μM | 24/48 h | suppresses cell proliferation in a dose-dependent manner and induces cell apoptosis | 24090995 | |
| MT-2 | Apoptosis Assay | 75-300 μM | 24/48 h | suppresses cell proliferation in a dose-dependent manner and induces cell apoptosis | 24090995 | |
| H460 | Apoptosis Assay | 100 nM | 24 h | enhances cell death co-treated with LY294002 | 24472538 | |
| A459 | Apoptosis Assay | 100 nM | 24 h | induces cell apoptosis co-treated with BEZ235 | 24472538 | |
| H460 | Apoptosis Assay | 100 nM | 24 h | induces cell apoptosis co-treated with BEZ235 | 24472538 | |
| GC | Growth Inhibition Assay | 50-125 μM | 72 h | attenuates the cell growth in a dose-dependent manner | 25774503 | |
| GH3 | Growth Inhibition Assay | 50-125 μM | 72 h | attenuates the cell growth in a dose-dependent manner | 25774503 | |
| BT474R | Function Assay | 50 μM | 10-60 d | inhibits STAT3 activity | 25327561 | |
| NCI-N87R | Function Assay | 50 μM | 10-60 d | inhibits STAT3 activity | 25327561 | |
| MDA-MB-468 | Function assay | 100 uM | 24 hrs | Inhibition of Stat3 activation in human MDA-MB-468 cells at 100 uM after 24 hrs | 17463090 | |
| MDA-MB-435 | Function assay | 100 uM | 24 hrs | Inhibition of Stat3 activation in human MDA-MB-435 cells at 100 uM after 24 hrs | 17463090 | |
| MDA-MB-231 | Function assay | 100 uM | 24 hrs | Inhibition of Stat3 activation in human MDA-MB-231 cells at 100 uM after 24 hrs | 17463090 | |
| NIH3T3 | Function assay | 100 uM | 24 hrs | Reduction of pTyr-705 Stat3 level in v-Src expressing mouse NIH3T3 cells at 100 uM after 24 hrs | 17463090 | |
| NIH3T3 | Growth inhibition assay | 100 uM | 4 days | Growth inhibition of mouse NIH3T3 cells expressing v-Src at 100 uM after 4 days by trypan blue exclusion assay | 17463090 | |
| MDA-MB-435 | Growth inhibition assay | 100 uM | 4 days | Growth inhibition of human MDA-MB-435 cells expressing v-Src at 100 uM after 4 days by trypan blue exclusion assay | 17463090 | |
| MDA-MB-231 | Growth inhibition assay | 100 uM | 4 days | Growth inhibition of human MDA-MB-231 cells expressing v-Src at 100 uM after 4 days by trypan blue exclusion assay | 17463090 | |
| MDA-MB-468 | Growth inhibition assay | 100 uM | 4 days | Growth inhibition of human MDA-MB-468 cells expressing v-Src at 100 uM after 4 days by trypan blue exclusion assay | 17463090 | |
| NIH3T3 | Growth inhibition assay | 100 uM | Growth inhibition of mouse NIH3T3 cells expressing v-Ras at 100 uM for every 3 days by soft-agar colony-formation assay | 17463090 | ||
| MDA-MB-435 | Apoptosis assay | 30 to 100 uM | 48 hrs | Induction of apoptosis in human MDA-MB-435 cells expressing active Stat3 at 30 to 100 uM after 48 hrs | 17463090 | |
| MDA-MB-231 | Apoptosis assay | 100 uM | 24 hrs | Reduction of apoptosis in Stat3 transfected human MDA-MB-231 cells at 100 uM after 24 hrs | 17463090 | |
| MDA-MB-231 | Function assay | 100 uM | 48 hrs | Reduction of cyclin D1 gene expression in human MDA-MB-231 cells at 100 uM after 48 hrs | 17463090 | |
| MDA-MB-231 | Apoptosis assay | 100 uM | 24 hrs | Induction of apoptosis in Stat3 SH2 domain transfected human MDA-MB-231 cells at 100 uM after 24 hrs | 17463090 | |
| MDA-MB-231 | Apoptosis assay | 100 uM | 24 hrs | Induction of apoptosis in Stat3C transfected human MDA-MB-231 cells at 100 uM after 24 hrs | 17463090 | |
| NIH3T3 | Function assay | 100 uM | 48 hrs | Reduction of cyclin D1 gene expression in v-Src transfected mouse NIH3T3 cells at 100 uM after 48 hrs | 17463090 | |
| NIH3T3 | Function assay | 100 uM | 48 hrs | Reduction of Bcl-xL gene expression in v-Src transfected mouse NIH3T3 cells at 100 uM after 48 hrs | 17463090 | |
| NIH3T3 | Function assay | 100 uM | 48 hrs | Reduction of survivin gene expression in v-Src transfected mouse NIH3T3 cells at 100 uM after 48 hrs | 17463090 | |
| MDA-MB-231 | Function assay | 100 uM | 48 hrs | Reduction of Bcl-xL gene expression in human MDA-MB-231 cells at 100 uM after 48 hrs | 17463090 | |
| MDA-MB-231 | Function assay | 100 uM | 48 hrs | Reduction of survivin gene expression in human MDA-MB-231 cells at 100 uM after 48 hrs | 17463090 | |
| MDA-MB-231 | Antitumor assay | 5 mg/kg | 2 weeks | Antitumor activity against human MDA-MB-231 cells expressing active Stat3 xenografted in mouse at 5 mg/kg, iv for every 3 days for 2 weeks | 17463090 | |
| NIH3T3 | Growth inhibition assay | 100 uM | Growth inhibition of mouse NIH3T3 cells expressing v-Src at 100 uM for every 3 days by soft-agar colony-formation assay | 17463090 | ||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Biological Activity
| Description | NSC 74859 (S3I-201) shows potent inhibition of STAT3 DNA-binding activity with IC50 of 86 μM in cell-free assays, and low activity towards STAT1 and STAT5. | ||
|---|---|---|---|
| Features | A chemical probe inhibitor of Stat3 activity. | ||
| Targets |
|
| In vitro | ||||
| In vitro | S3I-201 inhibits growth and induces apoptosis preferentially in tumor cells that contain persistently activated Stat3 by inhibiting Stat3·Stat3 complex formation and Stat3 DNA-binding and transcriptional activitie. Moreover, S3I-201 also inhibits the expression of the Stat3-regulated genes encoding cyclin D1, Bcl-xL, and survivin. [1] S3I-201 inhibits breast carcinoma MDA-MB-435, MDA-MB-453 and MDA-MB-231 cell lines with IC50 of 100 μM. In addition, the cells with impaired TGF-β signaling are four times as sensitive to the STAT3 inhibitor S3I-201. [2] A recent study shows that S3I-201 potentiates the antiproliferative effect in HepG2 and Huh-7 cells via the STAT3 signalling pathway. [3] |
|||
|---|---|---|---|---|
| Kinase Assay | In vitro Stat3 DNA-binding assay and EMSA analysis | |||
| Briefly, 100 mL of biotinyl-e-Ac-EPQpYEEIEL-OH (in 50 mM Tris/150 mM NaCl, pH 7.5) is added to each well of streptavidin-coated 96-well microtiter plates and incubated with shaking at 4 °C overnight. Then plates are rinsed with PBS/Tween 20 and then two times with 200 mL of BSA-T-PBS (0.2% BSA/0.1% Tween 20/PBS). Then 50 mL of Lck-SH2-GST fusion protein (6.4 ng/ml in BSA-T-PBS) is added to each well of the 96-well plate in the presence and absence of 50 mL of S3I-201 (for 30 and 100 mM final concentrations), and the plate is shaken at room temperature for 4 hours. After solutions are removed, each well is rinsed four times with BSA-T-PBS (200 mL), and 100 mL of polyclonal rabbit anti-GST antibody (100 ng/mL in BSA-T-PBS) is added to each well and incubated at 4 °C overnight. After washing with BSA-T-PBS, 100 mL of 200 ng/mL BSA-T-PBS horseradish peroxidase-conjugated mouse anti-rabbit antibody is added to each well and incubated for 45 minutes at room temperature. After four washing steps with BSA-T-PBS and three washing steps with PBS-T, 100 mL of peroxidase substrate is added to each well and incubated for 5-15 minutes. The peroxidase reaction is stopped by adding 100 mL of 1 M sulfuric acid solution, and absorbance is read at 450 nm with an ELISA plate rea | ||||
| Cell Research | Cell lines | MDA-MB-435, MDA-MB-453 and MDA-MB-231 cells lines | ||
| Concentrations | ~ 250 μM | |||
| Incubation Time | 72 hours | |||
| Method | The MTT assay is based on the conversion of the yellow tetrazolium salt MTT to purple formazan crystals by metabolically active cells. The MTT assay provides a quantitative determination of viable cells. Cells are seeded in 96-well microplates in complete culture medium in the absence or presence of increasing serial dosages of S3I-201 as indicated. At 72 hours after culture, the number of viable cells is measured by adding 100 μL/well of 2 mg/mL MTT solution. After 2 hours, the medium is removed. The absorbance is read at 590 nm with an enzyme-linked immunosorbent assay reader. Each treatment point is performed in 10 wells or sextuplicate. |
|||
| Experimental Result Images | Methods | Biomarkers | Images | PMID |
| Western blot | STAT3 / p-STAT3 / Cyclin D1 / Bcl-2 / Nanog / OCT4 / ALDH1 / CD44 PD-L1 |
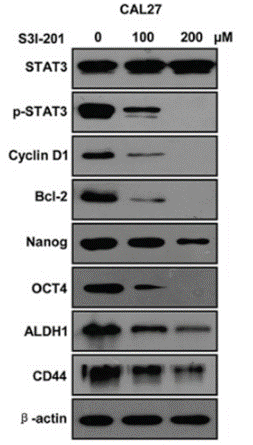
|
26556875 | |
| Immunofluorescence | p-STAT3 Oct4 / Twist |
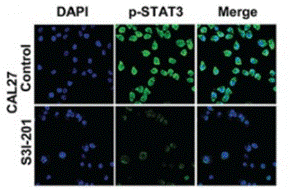
|
26556875 | |
| Growth inhibition assay | Cell viability |
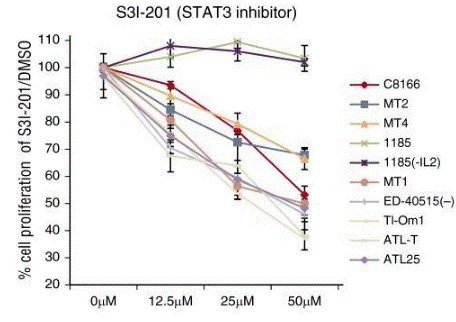
|
26813676 | |
| In Vivo | ||
| In vivo | S3I-201 (5 mg/kg, i.v. every 2 or every 3 days) shows the antitumor efficacy in mouse models with human breast tumor xenografts that harbor constitutively active Stat3. [1] S3I-201 treatment reduces Varicella-zoster virus (VZV) replication on the basis of the bioluminescence signal and the number of positive skin xenografts compared with DMSO-treated mice by inhibiting STAT3 phosphorylation. [4] |
|
|---|---|---|
| Animal Research | Animal Models | Human breast cancer MDA-MB-231 cells are injected s.c. into the left flank of athymic nu/nu mice. |
| Dosages | ≤5 mg/kg | |
| Administration | Administered via i.v. | |
|
Chemical Information & Solubility
| Molecular Weight | 365.36 | Formula | C16H15NO7S |
| CAS No. | 501919-59-1 | SDF | Download NSC 74859 (S3I-201) SDF |
| Smiles | CC1=CC=C(C=C1)S(=O)(=O)OCC(=O)NC2=CC(=C(C=C2)C(=O)O)O | ||
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 73 mg/mL ( (199.8 mM) Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Water : Insoluble Ethanol : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
|||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field
Tags: buy NSC 74859 (S3I-201) | NSC 74859 (S3I-201) supplier | purchase NSC 74859 (S3I-201) | NSC 74859 (S3I-201) cost | NSC 74859 (S3I-201) manufacturer | order NSC 74859 (S3I-201) | NSC 74859 (S3I-201) distributor