Topoisomerases can manage DNA's topological state in the nuclear to facilitate the replication, recombination, transcription and repair of DNA by separating the two strands of the helix temporarily. There are 2 types of DNA topoisomerases which are type I topoisomerase and type II topoisomerase. [show the full text]
Topoisomerase inhibitors
Other DNA Damage/DNA Repair Related Inhibitors
| Cat.No. | Product Name | Information | Product Use Citations | Product Validations |
|---|---|---|---|---|
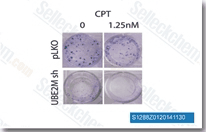
| S1288 | Camptothecin (CPT) | Camptothecin (CPT) is a specific inhibitor of DNA topoisomerase I (Topo I) with IC50 of 0.68 μM in a cell-free assay. Camptothecin induces apoptosis in cancer cells via microRNA-125b-mediated mitochondrial pathways. Phase 2. |
|
|
| E2516 | Doxorubicin | Adriamycin (Doxorubicin, Hydroxydaunorubicin), a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent, inhibits topoisomerase II with an IC50 of 2.67 μM, thus stopping DNA replication, and induces apoptosis. |
|
|
| S1198 | Irinotecan (CPT-11) | Irinotecan is a topoisomerase I inhibitor for LoVo cells and HT-29 cells with IC50 of 15.8 μM and 5.17 μM, respectively. |
|

|
| S1208 | Doxorubicin (Adriamycin) Hydrochloride | Doxorubicin (DOX) HCl is an antibiotic agent that inhibits human DNA topoisomerase II with IC50 of 2.67 μM. Doxorubicin reduces basal phosphorylation of AMPK. Doxorubicin is used in the concomitant treatment of HIV-infected patients but is found to be at high risk of HBV reactivation.This product may precipitate when dissolved in PBS solution. It is recommended to prepare the stock solution in pure water and dilute with either pure water or saline to obtain the working solution.Doxorubicin (Adriamycin) HCl can be used to induce animal models of kidney disease. |
|

|
| S1225 | Etoposide | Etoposide is a semisynthetic derivative of podophyllotoxin, which inhibits DNA synthesis via topoisomerase II inhibition activity which enhances double-strand and single-strand cleavage of DNA and reversibly inhibits repair by topoisomerase II binding. Etoposide induces autophagy, mitophagy and apoptosis. |
|

|
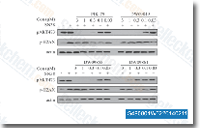
| S4908 | SN-38 | SN-38 (NK012) is an active metabolite of CPT-11, inhibits DNA topoisomerase I, DNA synthesis and causes frequent DNA single-strand breaks. SN-38 induces autophagy. |
|
|
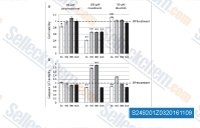
| S2492 | Novobiocin (Albamycin) Sodium | Novobiocin Sodium (NSC 2382, Albamycin, Cathomycin) is an aminocoumarin antibiotic that targets bacterial DNA gyrase (TopoIV), used to treat susceptible gram positive bacteria. |
|
|
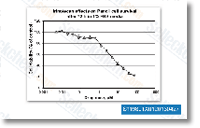
| S2217 | Irinotecan Hydrochloride Trihydrate | Irinotecan HCl Trihydrate is a hydrochloride trihydrate of irinotecan (Camptosar, Campto, CPT-11) which is a topoisomerase I inhibitor with IC50 of 15.8 and 5.17 μM for LoVo cells and HT-29 cells, respectively. |
|
|
| S1228 | Idarubicin HCl | Idarubicin HCl (4-demethoxydaunorubicin (NSC256439, 4-DMDR) HCl) is a hydrochloride salt form of Idarubicin which is an anthracycline antibiotic and a DNA topoisomerase II (topo II) inhibitor for MCF-7 cells with IC50 of 3.3 ng/mL in a cell-free assay. Idarubicin induces mTOR-dependent cytotoxic autophagy. |
|

|
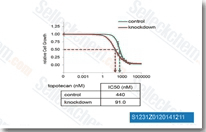
| S1231 | Topotecan HCl | Topotecan HCl is a topoisomerase I inhibitor for MCF-7 Luc cells and DU-145 Luc cells with IC50 of 13 nM and 2 nM in cell-free assays, respectively. This compound induces autophagy and apoptosis. |
|
|
Signaling Pathway Map
DNA topoisomerases are nuclear enzymes that play a critical role in DNA transcription and replication events for the efficient creation and compaction of two identical genomes in two daughter cells. There are at least five different topoisomerase that have been found in higher eukaryotes that can be grouped into two categories: (1) type I family, includes topoisomerases I, IIIα, IIIβ, and (2) type II family, includes topoisomerases IIα and IIβ.[1][2]
Type I enzymes, which do not require ATP, cleave one DNA strand at a time to achieve DNA strand relaxation. More specifically, among type I family constituents, topoisomerase I-mediated DNA strand scission involves a nucleophilic attack by the active site tyrosine OH group on the DNA phosphodiester bond at the site of cleavage. Such an attack results in the breakage of the DNA phosphodiester backbone and the creation of a phosphotyrosine bond between the enzyme and DNA. This covalent binary complex DNA-topoisomerase I, the so-called cleavable complex, is typically only an intermediate. Relaxation via passage (swivel movement) of the broken DNA strand around the unbroken strand is followed by reformation of the phosphodiester backbone as a result of relegation, with concomitant release of topoisomerase I and enzyme turnover.[1][2]
In contrast, type II enzymes which are typically ATP-dependent are able to perform double strand cuts that relieve superhelical twists, intramolecular DNA knots, and intermolecular tangles for chromosomal segregation to produce a DNA-linked protein gate through which another intact duplex can pass. It should be emphasized that the enzyme shows strong preference for supercoiled DNA versus relaxed molecules. More specifically, with topoisomerase II enzymes it is observed that DNA cleavage occurs at preferred sequences within its recognition/binding sites, but there is not clear specificity.[1][2]
In either case, both types of topoisomerases cleave DNA at the phosphodiester backbone by nucleophilic attack from a catalytic tyrosine residue which becomes linked to the phosphate end (P-Y) of the DNA break. The reactions of both types of topoisomerases are highly reversible and leave the DNA sequence unchanged following topoisomerization.[1][2]
While both topoisomerases can relax supercoiled DNA, only topoisomerase II can decatenate DNA molecules. Interestingly, throughout the cell cycle topoisomerase I and topoisomerase IIβ do not change in concentration, meanwhile topoisomerase IIα protein level are noted to fluctuate in relation to the proliferative stage and cell cycle position. In particular, topoisomerase IIα mRNA peak in late S and G2/M several-fold over (typically more than 10 times) the amount observed in G1 cells. The high levels of topoisomerase IIα during the final stages of DNA replication is intended to assist with chromosome untangling, condensation and mitotic segregation events. Consequently, cancerous cells are noted to have high topoisomerase IIα activity, and these findings have prompted researchers to develop new anti-cancer agents that specifically target to poisomerase II.[1][2]
In general, topoisomerase I or topoisomerase II-directed anti-cancer agents are able to interfere with at least one step of the catalytic cycle of the enzyme. Among the topoisomerase I inhibitor class of compounds, Camptothecin (CPT) and its derivatives – a pentacyclic alkaloid formerly isolated as a natural extract from the Chinese tree Camptoteca acuminate – are effective at selectively targeting topoisomerase I by trapping its catalytic intermediate during the topoisomerase I-DNA reaction. Agents that effectively target topoisomerase II include the Anthracyclines (i.e. Adriamycin and Daunorubicin, 9 and 10), Epipodophyllotoxins (i.e. Etoposide and Teniposide 11 and 12), Antracendedione (i.e. Mitoxantrone, 13) and Aminoacrideines (i.e. m-AMSA). The compounds are successful at stabilizing the short-lived covalent complexes between topoisomerase II and DNA. The anti-cancer agents convert the topoisomerase II enzymes into DNA-cleaving toxins which are currently are area of research interest.[2]





































