
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
Carfilzomib (PR-171) Proteasome inhibitor
Cat.No.S2853
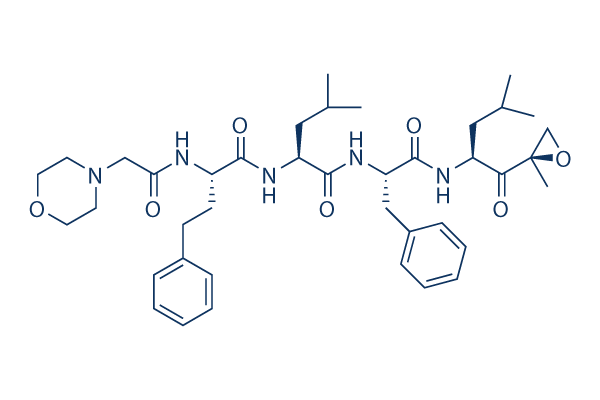
Chemical Structure
Molecular Weight: 719.91
Quality Control
Batch:
Purity:
99.75%
99.75
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| MM.1S | Growth Inhibition Assay | 0-100 nM | 48 h | IC50 = 10 nM | 25312543 | |
| NCI-H929 | Growth Inhibition Assay | 0-100 nM | 48 h | IC50 = 14 nM | 25312543 | |
| SUDHL16 | Apoptosis Asssay | 2.5–3.5 nM | 48 h | enhances the cell death co-treatment with ACY1215 | 25239935 | |
| SUDHL14 | Apoptosis Asssay | 2.5–3.5 nM | 48 h | enhances the cell death co-treatment with ACY1215 | 25239935 | |
| U2932 | Apoptosis Asssay | 2.5–3.5 nM | 48 h | enhances the cell death co-treatment with ACY1215 | 25239935 | |
| P-UMSCC-1 | Growth Inhibition Assay | IC50=11.2 nM | 24915039 | |||
| R-UMSCC-1 | Growth Inhibition Assay | IC50=2294 nM | 24915039 | |||
| P-Cal33 | Growth Inhibition Assay | IC50=17.3 nM | 24915039 | |||
| R-Cal33 | Growth Inhibition Assay | IC50=1112 nM | 24915039 | |||
| Jurkat | Growth Inhibition Assay | 1-11nM | 48 h | inhibits the cell proliferation co-treatment with vorinostat | 24801128 | |
| Jurkat | Apoptosis Asssay | 8 nM | 24/48 h | induces apoptosis, caspase activation, and PARP cleavage co-treatment with vorinostat | 24801128 | |
| UMSCC-22A | Apoptosis Asssay | 200 nM | 24 h | induce the cell apoptosis co-treatment with ONX 0912 | 22929803 | |
| UMSCC-22B | Apoptosis Asssay | 200 nM | 24 h | induce the cell apoptosis co-treatment with ONX 0912 | 22929803 | |
| 1483 | Apoptosis Asssay | 200 nM | 24 h | induce the cell apoptosis co-treatment with ONX 0912 | 22929803 | |
| UMSCC-1 | Apoptosis Asssay | 200 nM | 24 h | induce the cell apoptosis co-treatment with ONX 0912 | 22929803 | |
| UMSCC-22A | Growth Inhibition Assay | IC50=38.7 ± 1.0 nM | 22929803 | |||
| UMSCC-22B | Growth Inhibition Assay | IC50=30.7 ± 9.3 nM | 22929803 | |||
| 1483 | Growth Inhibition Assay | IC50=50.5 ± 11.9 nM | 22929803 | |||
| UMSCC-1 | Growth Inhibition Assay | IC50=34.6 ± 2.6 nM | 22929803 | |||
| Cal33 | Growth Inhibition Assay | IC50=49.3 ± 8.9 nM | 22929803 | |||
| PCI-15A | Growth Inhibition Assay | IC50=70.4 ± 22.6 nM | 22929803 | |||
| PCI-15B | Growth Inhibition Assay | IC50=39.5 ± 11.0 nM | 22929803 | |||
| OSC-19 | Growth Inhibition Assay | IC50=18.3 ± 4.2 nM | 22929803 | |||
| SUDHL16 | Apoptosis Asssay | 2.0-4.0 nM | 48 h | induces cell death co-treatment with obatoclax | 22411899 | |
| SUDHL16 | Function Assay | 2.5 nM | 24 h | activates JNK, inactivates AKT, up-regulates Noxa, and induces γH2A.X co-treatment with obatoclax | 22411899 | |
| Granta | Growth Inhibition Assay | 0-4 nM | 48 h | induce cell death co-treatment with HADCIs | 21750224 | |
| SUDHL16 | Growth Inhibition Assay | 1-4 nM | 36 h | induce cell death co-treatment with HADCIs | 20233973 | |
| MOLT4 | Function assay | 1 hr | Inhibition of chymotrypsin-like activity of 20S proteasome in human MOLT4 cells after 1 hr by CellTiter-Glo luminescent assay, IC50 = 0.0051 μM. | 19348473 | ||
| MESSA | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MESSA cells assessed as cell viability after 72 hrs by CellTiter-Glo luminescent assay, IC50 = 0.018 μM. | 19348473 | ||
| MESSA | Cytotoxicity assay | 72 hrs | Cytotoxicity against multidrug resistance transporter expressing doxorubicin resistant human MESSA cells assessed as cell viability after 72 hrs by CellTiter-Glo luminescent assay, IC50 = 0.413 μM. | 19348473 | ||
| RPMI8226 | Cytotoxicity assay | 72 hrs | Cytotoxic activity against human RPMI8226 cells after 72 hrs by MTS assay, IC50 = 0.01319 μM. | 24767818 | ||
| NCI-H929 | Cytotoxicity assay | 72 hrs | Cytotoxic activity against human NCI-H929 cells after 72 hrs by MTS assay, IC50 = 0.02132 μM. | 24767818 | ||
| CCRF-CEM | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CCRF-CEM cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0061 μM. | 26231162 | ||
| RPMI8266 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human RPMI8266 cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0139 μM. | 26231162 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0193 μM. | 26231162 | ||
| A431 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A431 cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0238 μM. | 26231162 | ||
| TOV21G | Antiproliferative assay | 72 hrs | Antiproliferative activity against human TOV21G cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0238 μM. | 26231162 | ||
| RKO | Antiproliferative assay | 72 hrs | Antiproliferative activity against human RKO cells after 72 hrs by oxyluciferin luminescence assay, IC50 = 0.0271 μM. | 26231162 | ||
| MM1S | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MM1S cells measured after 72 hrs by MTS assay, IC50 = 0.0015 μM. | 27765408 | ||
| RPMI8226 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human RPMI8226 cells measured after 72 hrs by MTS assay, IC50 = 0.0132 μM. | 27765408 | ||
| LCL | Cytotoxicity assay | 48 hrs | Cytotoxicity against human LCL cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.03 μM. | 27994734 | ||
| RD-ES | Cytotoxicity assay | 48 hrs | Cytotoxicity against human RD-ES cells harboring p53 mutant assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.043 μM. | 27994734 | ||
| U266 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human U266 cells harboring mutant p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.06 μM. | 27994734 | ||
| WE68 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human WE68 cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.08 μM. | 27994734 | ||
| IMR90 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human IMR90 cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.13 μM. | 27994734 | ||
| MCF10A | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MCF10A cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.32 μM. | 27994734 | ||
| SKOV3 | Cytotoxicity assay | 48 hrs | Cytotoxicity against p53 deficient human SKOV3 cells assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.32 μM. | 27994734 | ||
| MDA-MB-468 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MDA-MB-468 cells harboring mutant p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.33 μM. | 27994734 | ||
| HNDF | Cytotoxicity assay | 48 hrs | Cytotoxicity against HNDF cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.35 μM. | 27994734 | ||
| KGN | Cytotoxicity assay | 48 hrs | Cytotoxicity against human KGN cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 0.45 μM. | 27994734 | ||
| MCF7 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MCF7 cells harboring wild type p53 assessed as reduction in cell viability after 48 hrs by 7AAD-staining based FACS analysis, LD50 = 4.5 μM. | 27994734 | ||
| MCF7 | Function assay | 35 nM | 4 hrs | Inhibition of 26S proteasome in human MCF7 cells assessed as accumulation of high molecular weight polyubiquitin-conjugated proteins at 35 nM after 4 hrs by Western blot analysis | 27994734 | |
| MDA-MB-468 | Function assay | 35 nM | 4 hrs | Inhibition of 26S proteasome in human MDA-MB-468 cells assessed as accumulation of high molecular weight polyubiquitin-conjugated proteins at 35 nM after 4 hrs by Western blot analysis | 27994734 | |
| MM1S | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MM1S cells measured after 72 hrs by MTS assay, IC50 = 0.0015 μM. | 28027531 | ||
| RPMI8226 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human RPMI8226 cells measured after 72 hrs by MTS assay, IC50 = 0.0132 μM. | 28027531 | ||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for TC32 cells | 29435139 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for U-2 OS cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for DAOY cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Saos-2 cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-12 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for BT-12 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for LAN-5 cells | 29435139 | |||
| fibroblast cells | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for control Hh wild type fibroblast cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for NB-EBc1 cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-SH cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh41 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for A673 cells) | 29435139 | |||
| Rh30 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh30 cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for BT-37 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for MG 63 (6-TG R) cells | 29435139 | |||
| Rh30 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh30 cells | 29435139 | |||
| fibroblast cells | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for control Hh wild type fibroblast cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for OHS-50 cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for SK-N-SH cells | 29435139 | |||
| Daoy | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for Daoy cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D caspase screen for TC32 cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for TC32 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for MG 63 (6-TG R) cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh18 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-MC cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SJ-GBM2 cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for TC32 cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh18 cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Saos-2 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for SJ-GBM2 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for RD cells | 29435139 | |||
| ANBL-6 | Function assay | Inhibition of 20S proteasome activity in human ANBL-6 cells, IC50 = 0.01 μM. | 29652143 | |||
| MCF7 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MCF7 cells after 48 hrs by MTT assay, IC50 = 0.0041 μM. | 30165344 | ||
| MDA-MB-231 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MDA-MB-231 cells after 48 hrs by MTT assay, IC50 = 0.0044 μM. | 30165344 | ||
| RPMI8226 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human RPMI8226 cells after 48 hrs by MTT assay, IC50 = 0.0067 μM. | 30165344 | ||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 719.91 | Formula | C40H57N5O7 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 868540-17-4 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC(C)CC(C(=O)C1(CO1)C)NC(=O)C(CC2=CC=CC=C2)NC(=O)C(CC(C)C)NC(=O)C(CCC3=CC=CC=C3)NC(=O)CN4CCOCC4 | ||
Solubility
|
In vitro |
DMSO
: 100 mg/mL
(138.9 mM)
Ethanol : 50 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki |
Proteasome [1]
(ANBL-6 cells) 5 nM
|
|---|---|
| In vitro |
Carfilzomib (PR-171) inhibits proliferation in a variety of cell lines and patient-derived neoplastic cells, including multiple myeloma, and induced intrinsic and extrinsic apoptotic signaling pathways and activation of c-Jun-N-terminal kinase (JNK). It reveals enhanced anti-MM activity, overcomes resistance to other agents, and acts synergistically with (Dex). This compound shows preferential in vitro inhibitory potency against the ChT-L activity in the β5 subunit, with over 80% inhibition at doses of 10 nM. Short exposure to low-dose Carfilzomib leads to preferential binding specificity for the β5 constitutive 20S proteasome and the β5i immunoproteasome subunits. Measurement of caspase activity in ANBL-6 cells pulsed with it reveals substantial increases in caspase-8, caspase-9, and caspase-3 activity after 8 hours, giving a 3.2-, 3.9- and 6.9-fold increase, respectively, over control cells after 8 hours. In carfilzomib pulse-treated cells, the mitochondrial membrane integrity is decreased to 41% (Q1 + Q2), compared with 75% in vehicle-treated control cells. [1] In another study, it has also shown preclinical effectiveness against hematological and solid malignancies. [2] It directly inhibits osteoclasts formation and bone resorption. [3] |
| Kinase Assay |
Enzyme-linked immunosorbent assay for subunit profiling of carfilzomib
|
|
ANBL-6 cells (2 × 106/well) are plated in 96-well plates and treated with Carfilzomib (PR-171) doses from 0.001 to 10 μM for 1 hour. Cells are then lysed (20 mM Tris-HCl, 0.5 mM EDTA), and cleared lysates are transferred to polymerase chain reaction (PCR) plates. A standard curve is generated using untreated ANBL-6 cell lysates starting at a concentration of 6 μg protein/μL. The active site probe [biotin-(CH2)4-Leu-Leu-Leu-epoxyketone; 20 μM] is added and incubated at room temperature for 1 hour. Cell lysates are then denatured by adding 1% sodium dodecyl sulfate (SDS) and heating to 100°C, followed by mixing with 20 μL per well streptavidin-sepharose high-performance beads in a 96-well multiscreen DV plate and incubated for 1 hour. These beads are then washed with enzyme-linked immunosorbent assay (ELISA) buffer (PBS, 1% bovine serum albumin, and 0.1% Tween-20), and incubated overnight at 4°C on a plate shaker with antibodies to proteasome subunits. Antibodies used included mouse monoclonal anti-β1, anti-β2, anti-β1i, and anti-β5i, goat polyclonal anti-β2i, and rabbit polyclonal anti-β5 (affinity-purified antiserum against KLH-CWIRVSSDNVADLHDKYS peptide). The beads are washed and incubated for 2 hours with horseradish peroxidase-conjugated secondary goat antirabbit, goat antimouse or rabbit antigoat antibodies. After washing, the beads are developed using the supersignal ELISA picochemiluminescence substrate. Luminescent detection is performed. Raw luminescence is converted to μg/mL by comparison with the standard curve and expressed as the % inhibition relative to vehicle control. Curve fits are generated using the following nonsigmoidal dose-response equation: Y = Bottom + (Top-Bottom)/(1 + 10̂((LogEC50 − X) × HillSlope)), where X is the logarithm of concentration, Y is the % inhibition, and EC50 is the dose of this compound showing 50% effect.
|
|
| In vivo |
Carfilzomib (PR-171) moderately reduces tumor growth in an in vivo xenograft model. It effectively decreases multiple myeloma cell viability following continual or transient treatment mimicking. This compound also increases trabecular bone volume, decreases bone resorption and enhances bone formation in non-tumor bearing mice. [3] |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | pERK / ERK / pSTAT5 / STAT5 / pPI3K / PI3K caspase-9 / caspase-8 c-PARP / PARP / caspase-3 Bcl-2 / Bcl-Xl / Mcl-1 / Bik / Bim / Bax / Bak Atg5 / Atg12 / Beclin-1 / LC3-II Noxa / Bik / Puma / Mcl-1 EGFR / HER2 / ER alpha / p-Akt(Ser473) / Akt / p-ERK / ERK / p53 BDP1 / HER2(Tyr1248) / HER2(Tyr1221/Tyr1222) / PARP1 / caspase-7 / p53 Mut HLA class I |
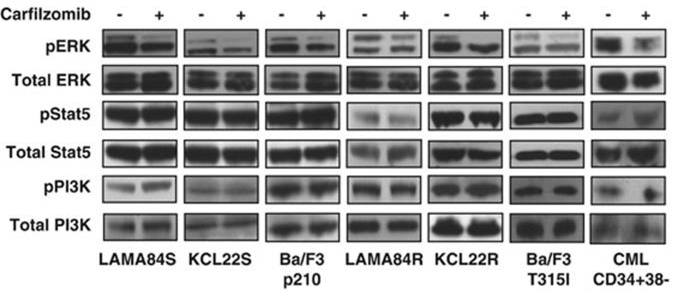
|
24590311 |
| Growth inhibition assay | Cell viability |
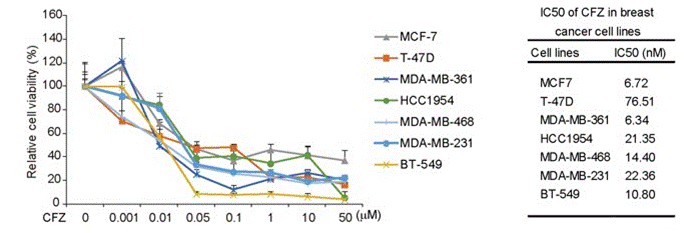
|
27655642 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT05552976 | Recruiting | Relapsed or Refractory Multiple Myeloma |
Bristol-Myers Squibb |
January 10 2023 | Phase 3 |
| NCT05675449 | Recruiting | Multiple Myeloma |
Pfizer |
December 14 2022 | Phase 1 |
| NCT05041933 | Unknown status | Hematological Diseases |
University Hospital Limoges |
September 15 2021 | -- |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
How should I prepare a solution of it for an ongoing in vivo study?
Answer:
It can be dissolved in 2% DMSO/30% PEG 300/dd H₂O at 10 mg/ml as a suspension, and can be dissolved in 2% DMSO/castor oil at 10 mg/ml as a clear solution.






































