
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
Sonidegib Hedgehog/Smoothened antagonist
Cat.No.S2151
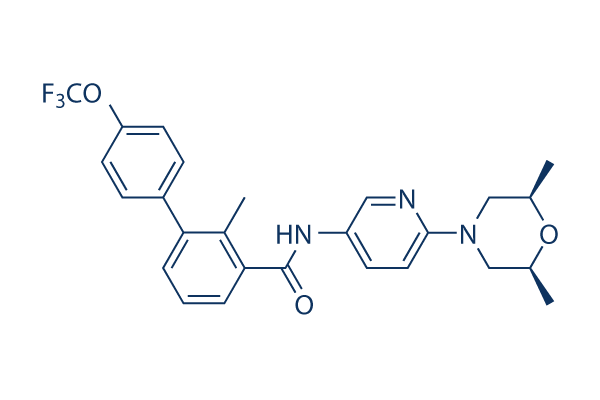
Chemical Structure
Molecular Weight: 485.5
Quality Control
Batch:
Purity:
99.90%
99.90
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| A2780ip2 | Cytoxicity assay | ~10 μM | IC50=12 μM | 22553355 | ||
| A2780cp20 | Cytoxicity assay | ~10 μM | IC50=7.5 μM | 22553355 | ||
| SKOV3ip1 | Cytoxicity assay | ~10 μM | IC50=24 μM | 22553355 | ||
| SKOV3TRip2 | Cytoxicity assay | ~10 μM | IC50=12 μM | 22553355 | ||
| HeyA8 | Cytoxicity assay | ~10 μM | IC50=18 μM | 22553355 | ||
| HeyA8MDR | Cytoxicity assay | ~10 μM | IC50=8 μM | 22553355 | ||
| OS5 | Growth inhibitory assay | ~5 μM | reduces the proliferation | 23243595 | ||
| OS18 | Growth inhibitory assay | ~5 μM | reduces the proliferation | 23243595 | ||
| Glioblastoma initiating cells | Cytoxicity assay | ~10 μM | Inhibits Cell Viability | 23482671 | ||
| Glioblastoma initiating cells | Function assay | ~10 μM | inhibits neurosphere formation | 23482671 | ||
| Glioblastoma initiating cells | Cytoxicity assay | ~10 μM | induces apoptosis | 23482671 | ||
| Glioblastoma initiating cells | Function assay | ~10 μM | downregulates the SHH signaling pathway | 23482671 | ||
| Glioblastoma initiating cells | Function assay | ~10 μM | Inhibits the Expression of Genes Involved in Maintaining Pluripotency | 23482671 | ||
| Glioblastoma initiating cells | Function assay | ~10 μM | Inhibits Motility, Invasion, and Migration | 23482671 | ||
| LOX IMVI | Function assay | 10 μM | DMSO | inhibits Hedgehog-GLI pathway | 23935925 | |
| UACC 257 | Function assay | 10 μM | DMSO | inhibits Hedgehog-GLI pathway | 23935925 | |
| LOX IMVI | Function assay | 10 μM | DMSO | induces G1 cell cycle arrest | 23935925 | |
| UACC 257 | Function assay | 10 μM | DMSO | induces G1 cell cycle arrest | 23935925 | |
| LOX IMVI | Cytoxicity assay | 10 μM | DMSO | decreases tumor cell viability | 23935925 | |
| UACC 257 | Cytoxicity assay | 10 μM | DMSO | decreases tumor cell viability | 23935925 | |
| LOX IMVI | Apoptosis assay | 10 μM | DMSO | induces apoptosis | 23935925 | |
| UACC 257 | Apoptosis assay | 10 μM | DMSO | induces apoptosis | 23935925 | |
| ACHN | Growth inhibitory assay | ~5 μM | DMSO | IC50=2-3 μM | 25093491 | |
| 769-P | Growth inhibitory assay | ~5 μM | DMSO | IC50=2-3 μM | 25093491 | |
| 786-O | Growth inhibitory assay | ~5 μM | DMSO | IC50=2-3 μM | 25093491 | |
| 786-O SuR | Growth inhibitory assay | ~5 μM | DMSO | IC50=2-3 μM | 25093491 | |
| SP53 | Function assay | 30 μM | DMSO | inhibits cell adhesion and migration | 26885608 | |
| SP53 | Function assay | 30 μM | DMSO | inhibits the VLA4-mediated FAK signaling pathway | 26885608 | |
| HS5 | Function assay | 30 μM | DMSO | inhibits cell adhesion and migration | 26885608 | |
| HS27a | Function assay | 30 μM | DMSO | inhibits cell adhesion and migration | 26885608 | |
| SP53 | Cytoxicity assay | 30 μM | DMSO | induces autophagy | 26885608 | |
| Jeko | Cytoxicity assay | 30 μM | DMSO | induces autophagy | 26885608 | |
| U2OS | Function assay | 2 hrs | Displacement of [3H]cyclopamine from wild type Smo expressed in U2OS cells after 2 hrs by scintillation counting, Ki = 0.006 μM. | 23063522 | ||
| NIH/3T3 | Function assay | Inhibition of hedgehog signaling pathway in mouse NIH/3T3 cells expressing wild type Smo assessed as reduction in Gli mRNA expression by RT-PCR method, IC50 = 0.0044 μM. | 27810591 | |||
| NIH/3T3 | Function assay | Inhibition of hedgehog signaling pathway in mouse NIH/3T3 cells harboring Smo D477H mutant assessed as reduction in Gli mRNA expression by RT-PCR method, IC50 = 0.0227 μM. | 27810591 | |||
| NIH/3T3 | Function assay | 24 hrs | Inhibition of hedgehog signaling pathway in mouse NIH/3T3 cells measured after 24 hrs by Gli-dual luciferase reporter gene assay, IC50 = 0.006 μM. | 27810591 | ||
| TM3 | Function assay | 48 hrs | Inhibition of Hh signaling pathway in mouse TM3 cells assessed as downregulation of Gli1 gene expression after 48 hrs by luciferase reporter gene assay, EC50 = 0.0012 μM. | 26976215 | ||
| NIH3T3 | Function assay | 48 hrs | Inhibition of SHH signaling pathway in mouse NIH3T3 cells measured after 48 hrs by Gli-luciferase reporter assay, IC50 = 0.0055 μM. | 24176396 | ||
| NIH3T3 | Function assay | 48 hrs | Inhibition of Sonic-induced hedgehog signalling in mouse NIH3T3 cells after 48 hrs by Gli-luciferase reporter assay, IC50 = 0.0055 μM. | 26820554 | ||
| NIH/3T3 | Function assay | Inhibition of hedgehog signaling pathway expressed in mouse NIH/3T3 cells assessed as inhibition of hedgehog-induced Gli-2 accumulation at tip of primary cilia by DAPI staining based confocal microscopic analysis | 27810591 | |||
| NIH/3T3 | Function assay | Inhibition of hedgehog signaling pathway in mouse NIH/3T3 cells assessed as inhibition of hedgehog-induced Smo-EGFP ciliary translocation by DAPI staining based confocal microscopic analysis | 27810591 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for MG 63 (6-TG R) cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh41 cells | 29435139 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for U-2 OS cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-12 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Saos-2 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 485.5 | Formula | C26H26F3N3O3 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 956697-53-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | NVP-LDE225, Erismodegib | Smiles | CC1CN(CC(O1)C)C2=NC=C(C=C2)NC(=O)C3=CC=CC(=C3C)C4=CC=C(C=C4)OC(F)(F)F | ||
Solubility
|
In vitro |
DMSO
: 97 mg/mL
(199.79 mM)
Ethanol : 16 mg/mL Water : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki | |
|---|---|
| In vitro |
Sonidegib (Erismodegib, NVP-LDE225) inhibits TM3 luciferized cell line with 0.6 nM and 8 nM, at the presence of 1 nM and 25 nM Hh agonist Ag1.5, respectively. [1]
|
| In vivo |
Sonidegib (Erismodegib, NVP-LDE225) is highly bound to mouse, rat, and human plasma proteins (>99%) and moderately bound to dog and monkey plasma proteins (77 and 85%, respectively). LDE225 has high permeability (90.8% in man) in the PAMPA assay. LDE225 shows good oral bioavailability ranging from 69 to 102% in preclinical species when dosed in solution. LDE225 is a weak base with a measured pKa of 4.20 and exhibits relatively poor aqueous solubility. LDE225 demonstrates dose-related antitumor activity. At a dose of 5 mg/kg/day qd, LDE225 significantly inhibits tumor growth, corresponding to a T/C value of 33%. When dosed at 10 and 20 mg/kg/day qd, LDE225 gives rise to 51 and 83% regression, respectively. Gli1 mRNA inhibition correlates with tumor and plasma exposure of LDE225. LDE225 successfully penetrates the blood−brain barrier in tumor-bearing animals and results in tumor growth inhibition after 4 days of treatment. [1] LDE225 significantly reduces the tumor volume by 95.7% in Rip1-Tag2 mice. LDE225 prolongs survival in Rip1Tag2 mice. LDE225 decreases expression of stromal markers in the LDE225-treated mice. [2]
|
References |
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-p130CAS / p130CAS / p-FAK / FAK / p-Paxillin / Paxillin pp70S6K / P70s6k Gli-1 / Gli-2 / p-Akt / Akt Bcl2 / c-myc / Cyclin D1 / pERK / ERK / pJNK / JNK / Caspase 3 / Cleaved caspase 3 |
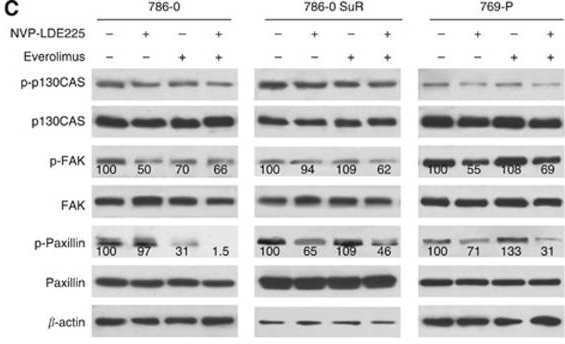
|
25093491 |
| Immunofluorescence | GLI1 |
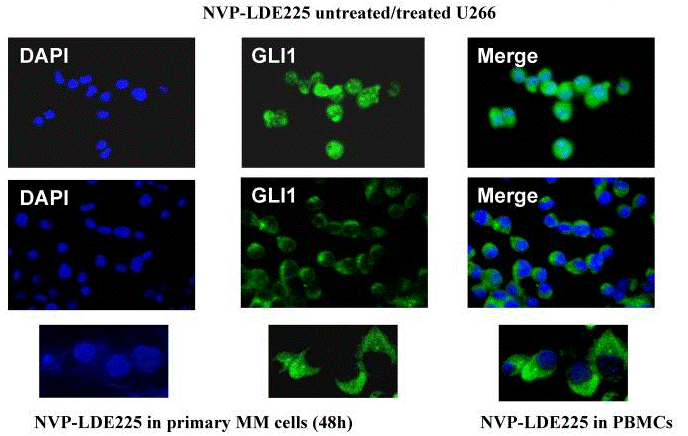
|
22821765 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT02358161 | Completed | Pancreatic Cancer |
Academisch Medisch Centrum - Universiteit van Amsterdam (AMC-UvA)|Novartis|Celgene Corporation |
September 2015 | Phase 1|Phase 2 |
| NCT02254551 | Terminated | Multiple Myeloma |
SCRI Development Innovations LLC|Novartis |
January 2015 | Phase 2 |
| NCT02138929 | Completed | Esophageal Cancer |
M.D. Anderson Cancer Center|Novartis|National Cancer Institute (NCI) |
November 10 2014 | Phase 1 |
| NCT02195973 | Completed | Recurrent Ovarian Cancer |
University of Alabama at Birmingham|Novartis Pharmaceuticals |
September 2014 | Phase 1 |
| NCT02027376 | Completed | Advanced Breast Cancer |
Spanish Breast Cancer Research Group|Novartis |
May 2014 | Phase 1 |
| NCT02111187 | Completed | Prostate Cancer |
Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins |
April 2014 | Phase 1 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































