Mitophagy inhibitors are a class of compounds that specifically block or interfere with the mitophagy process, a selective form of autophagy responsible for the degradation of damaged or dysfunctional mitochondria. As mitophagy plays a crucial role in maintaining cellular homeostasis, regulating energy metabolism, and mediating various pathological processes such as neurodegeneration, cancer, and cardiovascular diseases, the exploration of mitophagy inhibitors has become a frontier topic in biomedical research. These inhibitors not only serve as powerful tools to dissect the molecular mechanisms underlying mitophagy but also hold great potential for the development of novel therapeutic strategies targeting mitophagy-related disorders.
Mitophagy inducers/activators
| Cat.No. | Product Name | Information | Product Use Citations | Product Validations |
|---|---|---|---|---|
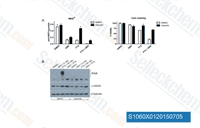
| S1060 | AZD2281 (Olaparib) | Olaparib (AZD2281, KU0059436) is a selective inhibitor of PARP1/2 with IC50 of 5 nM/1 nM in cell-free assays, 300-times less effective against tankyrase-1. Olaparib induces significant autophagy that is associated with mitophagy in cells with BRCA mutations. |
|
|
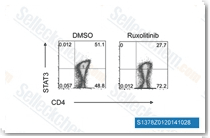
| S1378 | Ruxolitinib (INCB18424) | Ruxolitinib (INCB18424) is the first potent, selective, JAK1/2 inhibitor to enter the clinic with IC50 of 3.3 nM/2.8 nM in cell-free assays, >130-fold selectivity for JAK1/2 versus JAK3. This compound kills tumor cells through toxic mitophagy. It induces autophagy and enhances apoptosis. |
|
|
| S1208 | Doxorubicin (Adriamycin) Hydrochloride | Doxorubicin (DOX) HCl is an antibiotic agent that inhibits human DNA topoisomerase II with IC50 of 2.67 μM. Doxorubicin reduces basal phosphorylation of AMPK. Doxorubicin is used in the concomitant treatment of HIV-infected patients but is found to be at high risk of HBV reactivation.This product may precipitate when dissolved in PBS solution. It is recommended to prepare the stock solution in pure water and dilute with either pure water or saline to obtain the working solution.Doxorubicin (Adriamycin) HCl can be used to induce animal models of kidney disease. |
|

|
| S1225 | Etoposide | Etoposide is a semisynthetic derivative of podophyllotoxin, which inhibits DNA synthesis via topoisomerase II inhibition activity which enhances double-strand and single-strand cleavage of DNA and reversibly inhibits repair by topoisomerase II binding. Etoposide induces autophagy, mitophagy and apoptosis. |
|

|
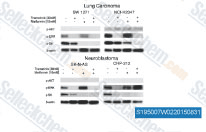
| S1950 | Metformin Hydrochloride | Metformin Hydrochloride (1,1-Dimethylbiguanide Hydrochloride) is a highly effective Antihyperglycemic Agent, which primarily decreases hyperglycemia in hepatocytes by suppressing hepatic gluconeogenesis (glucose production by the liver). It also promotes mitophagy in mononuclear cells and induces apoptosis of lung cancer cells through activating the JNK/p38 MAPK pathway and GADD153. |
|
|
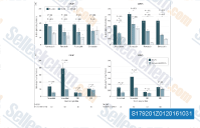
| S1792 | Simvastatin (MK-733) | Simvastatin is a competitive inhibitor of HMG-CoA reductase with Ki of 0.1-0.2 nM in cell-free assays. Simvastatin induces ferroptosis, mitophagy, autophagy and apoptosis. |
|
|
| S5243 | Ruxolitinib (INCB18424) Phosphate | Ruxolitinib Phosphate (INCB018424, INC424) is the phosphate salt form of Ruxolitinib. Ruxolitinib is the first potent, selective, JAK1/2 inhibitor to enter the clinic with IC50 of 3.3 nM/2.8 nM in cell-free assays, >130-fold selectivity for JAK1/2 versus JAK3. Ruxolitinib kills tumor cells through toxic mitophagy. Ruxolitinib induces autophagy and enhances apoptosis. |
|
|
| S1322 | Dexamethasone | Dexamethasone is a potent synthetic member of the glucocorticoid class of steroid drugs, and an interleukin receptor modulator that has anti-inflammatory and immunosuppressant effects. Dexamethasone induces autophagy and mitophagy. Dexamethasone is tested in hospitalized patients with COVID-19 and is found to have benefits for critically ill patients. |
|
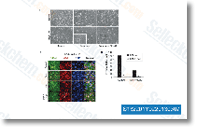
|
| S1759 | Pitavastatin (NK-104) Calcium | Pitavastatin calcium, a novel member of the medication class of statins, is a calcium salt formulation of pitavastatin which is a highly effective HMG-CoA reductase inhibitor. Pitavastatin Calcium attenuates AGEs-induced mitophagy via inhibition of ROS generation. Pitavastatin Calcium induces autophagy and apoptosis. |
|

|
| S2391 | Quercetin (Sophoretin) | Quercetin, a natural flavonoid present in vegetables, fruit and wine, is a stimulator of recombinant SIRT1 and also a PI3K inhibitor with IC50 of 2.4-5.4 μM. Quercetin induces mitophagy, apoptosis and protective autophagy. Phase 4. |
|
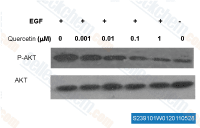
|
Classification of Mitophagy Inhibitors Based on Mitophagy Pathway
Mitophagy is regulated by multiple conserved pathways, and mitophagy inhibitors can be classified into different types according to the specific pathways they target. This classification is essential for understanding the specificity of inhibitors and their applicability in different research scenarios.
Inhibitors Targeting the PINK1-Parkin Pathway
The PINK1-Parkin pathway is the most well-characterized mitophagy pathway, and its dysregulation is closely associated with Parkinson's disease. Inhibitors targeting this pathway mainly act by blocking the activation of PINK1 or Parkin, or interfering with the downstream signaling events. For example, Cyclosporine A (CsA) has been shown to inhibit mitophagy by suppressing the interaction between Parkin and mitochondria, thereby preventing the ubiquitination of mitochondrial proteins that are essential for mitophagy initiation. Another inhibitor, N-acetylcysteine (NAC), exerts its inhibitory effect by scavenging reactive oxygen species (ROS), which are important triggers for PINK1 activation. Research on these inhibitors has greatly contributed to the understanding of the molecular details of the PINK1-Parkin pathway and its role in neurodegenerative diseases.
Inhibitors Targeting Non-PINK1-Parkin Pathways
In addition to the PINK1-Parkin pathway, mitophagy can also be induced through various non-canonical pathways, such as those mediated by BNIP3, NIX, and FUNDC1. Inhibitors targeting these pathways have unique mechanisms of action. For instance, 3-Methyladenine (3-MA), a classic autophagy inhibitor, can also block BNIP3-mediated mitophagy by inhibiting the class III PI3K complex, which is required for the formation of autophagosomes. Similarly, chloroquine (CQ) and hydroxychloroquine (HCQ) inhibit mitophagy by increasing the pH of lysosomes, thereby impairing the fusion between autophagosomes and lysosomes, a process that is essential for the degradation of mitophagosomes regardless of the initiating pathway. These non-specific inhibitors are widely used in research to study the general role of mitophagy in cellular processes, although their lack of pathway specificity needs to be considered when interpreting experimental results.
The Relationship Between Mitophagy Inhibitors and Autophagy Regulation
Mitophagy is a subset of autophagy, and thus, there is a close relationship between mitophagy inhibitors and autophagy regulators. Some mitophagy inhibitors are also general autophagy inhibitors, while others are specific to mitophagy, reflecting the differences in the regulatory mechanisms between general autophagy and selective mitophagy. Understanding this relationship is crucial for selecting appropriate inhibitors in research and avoiding off-target effects.
Overlap Between Mitophagy Inhibitors and General Autophagy Inhibitors
As mentioned earlier, 3-MA, CQ, and HCQ are typical examples of inhibitors that affect both autophagy and mitophagy. These compounds target the core machinery of autophagy, such as autophagosome formation and lysosomal function, which are shared by general autophagy and mitophagy. Therefore, their use in mitophagy research requires careful control experiments to distinguish the specific effects on mitophagy from those on general autophagy. For example, combining these inhibitors with mitophagy-specific markers can help confirm whether the observed effects are indeed due to the inhibition of mitophagy rather than general autophagy.
Specific Mitophagy Inhibitors and Their Independence from General Autophagy
With the deepening of research, an increasing number of mitophagy-specific inhibitors have been identified. These inhibitors selectively block mitophagy without significantly affecting general autophagy, making them ideal tools for studying the specific functions of mitophagy. For example, Mdivi-1, a small molecule inhibitor, specifically targets the mitochondrial fission protein Drp1, which is required for mitophagy but not for general autophagy. By inhibiting Drp1-mediated mitochondrial fission, Mdivi-1 blocks the segregation of damaged mitochondria from the mitochondrial network, thereby preventing their engulfment by autophagosomes. Studies using Mdivi-1 have revealed the specific role of mitophagy in regulating mitochondrial quality control and cellular survival under stress conditions.
Research Methods Involving Mitophagy Inhibitors: Parking and Fasting Models
Parking (Parkin-related) models and fasting models are commonly used in mitophagy research to induce mitophagy, and combining these models with mitophagy inhibitors can provide valuable insights into the physiological and pathological roles of mitophagy. These models simulate different physiological conditions, allowing researchers to study mitophagy in a more relevant biological context.
Mitophagy Inhibitors in Parking Models
Parking models are established by manipulating the expression or activity of Parkin, a key regulator of the PINK1-Parkin mitophagy pathway. These models are widely used to study the role of mitophagy in Parkinson's disease. For example, in Parkin-overexpressing cells or animal models, mitophagy is significantly enhanced. By treating these models with mitophagy inhibitors such as CsA or Mdivi-1, researchers can investigate the consequences of mitophagy inhibition on mitochondrial function, neuronal survival, and disease progression. These studies have provided important evidence for the involvement of mitophagy dysregulation in Parkinson's disease and have laid the foundation for the development of therapeutic strategies targeting mitophagy.
Mitophagy Inhibitors in Fasting Models
Fasting is a physiological stimulus that induces both general autophagy and mitophagy to provide energy for cells during nutrient deprivation. Fasting models are widely used to study the physiological functions of mitophagy. When mitophagy inhibitors are applied in fasting models, they can block the adaptive mitophagy response, leading to the accumulation of damaged mitochondria and impaired energy metabolism. For example, in fasting mice treated with CQ, the levels of mitophagy markers such as LC3-II/LC3-I ratio and p62 degradation are significantly reduced, and the mice show decreased tolerance to fasting due to impaired mitochondrial function. These studies have highlighted the importance of mitophagy in maintaining cellular homeostasis during nutrient stress and have provided insights into the role of mitophagy in metabolic disorders.
Detection of Mitophagy Markers in Research Using Mitophagy Inhibitors
The accurate detection of mitophagy markers is essential for evaluating the efficacy of mitophagy inhibitors and interpreting research results. Mitophagy markers can be divided into two categories: mitochondrial markers and autophagy markers, and their combination is often used to confirm mitophagy inhibition.
Mitochondrial Markers for Mitophagy Detection
Mitochondrial markers such as Tom20, COXIV, and Cytochrome c are commonly used to assess the degradation of mitochondria during mitophagy. When mitophagy is inhibited, the levels of these mitochondrial proteins increase due to the reduced degradation. For example, treatment with Mdivi-1 in cells under mitophagy-inducing conditions leads to the accumulation of Tom20 and COXIV, indicating the inhibition of mitophagy. In addition, mitochondrial DNA (mtDNA) content can also be used as a marker, as mitophagy inhibition results in the accumulation of mtDNA.
Autophagy-Mitophagy Fusion Markers
Autophagy-mitophagy fusion markers, such as the co-localization of LC3 (an autophagosome marker) with mitochondrial markers, are more specific for mitophagy. The use of immunofluorescence or confocal microscopy to detect the co-localization of LC3 and Tom20/COXIV can directly reflect the formation of mitophagosomes. When mitophagy is inhibited by inhibitors such as CQ or Mdivi-1, the co-localization of LC3 and mitochondrial markers is significantly reduced. In addition, the LC3-II/LC3-I ratio and the degradation of p62 (a substrate of autophagy) are also commonly used to assess autophagy and mitophagy activity, although they need to be combined with mitochondrial markers to confirm mitophagy-specific effects.
Interactions Between Mitophagy Inhibitors and Mitophagy Inducers in Research
Mitophagy inducers are compounds that promote mitophagy, and their combination with mitophagy inhibitors is often used in research to study the reversibility of mitophagy and to validate the specificity of inhibitors. This combination also provides a basis for the development of combination therapies targeting mitophagy-related diseases.
Validation of Mitophagy Inhibitor Specificity Using Inducers
Mitophagy inducers such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and antimycin A are commonly used to induce mitophagy by causing mitochondrial depolarization. When mitophagy inhibitors are applied together with these inducers, the inhibitory effect of the inhibitors can be evaluated by detecting mitophagy markers. For example, CCCP treatment increases the LC3-II/LC3-I ratio and reduces p62 levels, while co-treatment with Mdivi-1 or CQ reverses these effects, confirming the specificity of the inhibitors. This approach is widely used in the initial screening and validation of new mitophagy inhibitors.
Synergistic and Antagonistic Effects in Therapeutic Research
In therapeutic research targeting mitophagy-related diseases, the combination of mitophagy inhibitors and inducers may exhibit synergistic or antagonistic effects. For example, in cancer research, some cancer cells rely on enhanced mitophagy to survive under hypoxic conditions. The use of mitophagy inhibitors can block this adaptive response, making cancer cells more sensitive to chemotherapy or radiotherapy. On the other hand, mitophagy inducers may be beneficial in neurodegenerative diseases by promoting the clearance of damaged mitochondria. However, the inappropriate use of inhibitors or inducers may have adverse effects, highlighting the need for a comprehensive understanding of the role of mitophagy in specific diseases. The interaction between mitophagy inhibitors and inducers provides important clues for optimizing therapeutic strategies and avoiding potential side effects.
In conclusion, mitophagy inhibitors are essential tools in mitophagy research, providing valuable insights into the molecular mechanisms and physiological functions of mitophagy. The classification based on mitophagy pathways helps in selecting specific inhibitors for different research purposes, while the understanding of their relationship with autophagy regulators ensures the accuracy of research results. The application of mitophagy inhibitors in parking and fasting models simulates physiological and pathological conditions, facilitating the study of mitophagy in relevant biological contexts. The accurate detection of mitophagy markers is crucial for evaluating inhibitor efficacy, and the interaction with mitophagy inducers provides a basis for therapeutic development. Future research should focus on identifying more specific and potent mitophagy inhibitors, exploring their mechanisms of action in depth, and evaluating their therapeutic potential in various mitophagy-related diseases.





































