
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
Vemurafenib (PLX4032) B-Raf inhibitor
Cat.No.S1267
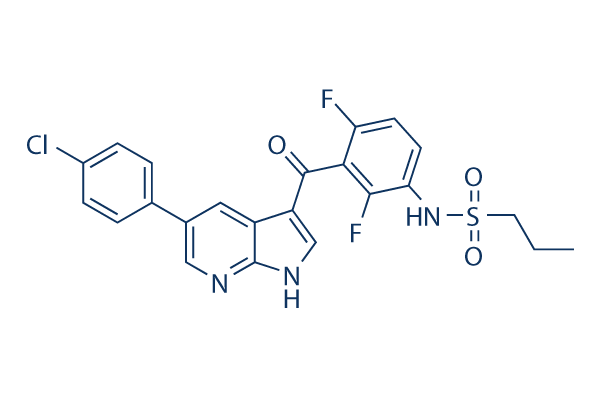
Chemical Structure
Molecular Weight: 489.92
Quality Control
Batch:
Purity:
99.98%
99.98
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| SKMEL19 | Function Assay | 6 μM | 48 h | DMSO | Triggers ER stress | 23362240 |
| VMM12 | Function Assay | 3 μM | 48 h | DMSO | Increases collagen synthesis and decreases IL-9 expression | 25989506 |
| C4 | Function Assay | 3 μM | 48 h | DMSO | Increases collagen synthesis and decreases IL-8 expression | 25989506 |
| Calu-6 | Function Assay | 1 μM | 1 h | DMSO | Activates MEK/ERK in cells with wild-type BRAF | 20179705 |
| PC | Growth Inhibition Assay | 96 h | EC50> 1000 nM | 19880792 | ||
| TPC-1 (RET/PTC1) | Growth Inhibition Assay | 96 h | EC50≥1000 nM | 19880792 | ||
| CAL62 (KRAS G12R) > 1000 > 1000 | Growth Inhibition Assay | 96 h | EC50> 1000 nM | 19880792 | ||
| HTH7 (NRAS Q61R) | Growth Inhibition Assay | 96 h | EC50≥ 1000 nM | 19880792 | ||
| C643 (HRAS G13R)≥ 500 | Growth Inhibition Assay | 96 h | EC50 ≥ 500 nM | 19880792 | ||
| BCPAP (BRAF WT/V600E) | Growth Inhibition Assay | 96 h | EC50=78 nM | 19880792 | ||
| BHT101 (BRAF WT/V600E) | Growth Inhibition Assay | 96 h | EC50=97 nM | 19880792 | ||
| SW1736 (BRAF WT/V600E) | Growth Inhibition Assay | 96 h | EC50=29 nM | 19880792 | ||
| 8505C (BRAF V600E/V600E) | Growth Inhibition Assay | 96 h | EC50=57 nM | 19880792 | ||
| ARO | Function Assay | 10 μM | 72 h | DMSO | Induces the reexpression of the NIS pump | 18458053 |
| A375 | Apoptosis Assay | 10 μM | DMSO | Promotes apoptotic death | 18458053 | |
| TPCI | Growth Inhibition Assay | 100 μM | 96 h | DMSO | IC50=10.77 μM | 18458053 |
| ARO | Growth Inhibition Assay | 100 μM | 96 h | DMSO | IC50=205 nM | 18458053 |
| NPA | Growth Inhibition Assay | 100 μM | 96 h | DMSO | IC50=26 nM | 18458053 |
| A375 | Growth Inhibition Assay | 100 μM | 96 h | DMSO | IC50=47 nM | 18458053 |
| UKF-NB-3 (ABCB1) | Function Assay | 1.25 µM | 2 h | DMSO | Enhances accumulation of the fluorescent ABCB1 substrate rhodamine 123 | 24735766 |
| UKF-NB-3 | Function Assay | 1.25 µM | 2 h | DMSO | Significantly affects on accumulation of the fluorescent ABCB1 substrate rhodamine 123 | 24735766 |
| A375 (BRAFV600E) | Function Assay | 8 h | DMSO | Increases intracellular ROS and NO levels | 25363644 | |
| A375P | Antiproliferative assay | Antiproliferative activity against human A375P cells, IC50 = 0.254 μM. | 22460030 | |||
| A375 | Antiproliferative assay | Antiproliferative activity against human A375 cells expressing B-Raf V600E mutant and wild type Ras, IC50 = 0.31 μM. | 22808911 | |||
| A375P | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A375P cells after 48 hrs by MTT assay, IC50 = 0.25 μM. | 24128410 | ||
| A375 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A375 cells after 72 hrs by MTT assay, IC50 = 0.18 μM. | 24215818 | ||
| SK-MEL-28 | Cytotoxicity assay | Cytotoxicity against human SK-MEL-28 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M229 | Cytotoxicity assay | Cytotoxicity against human M229 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M263 | Cytotoxicity assay | Cytotoxicity against human M263 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M321 | Cytotoxicity assay | Cytotoxicity against human M321 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M238 | Cytotoxicity assay | Cytotoxicity against human M238 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M262 | Cytotoxicity assay | Cytotoxicity against human M262 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M249 | Cytotoxicity assay | Cytotoxicity against human M249 cells expressing B-raf V600E mutant, IC50 = 0.1 μM. | 24471466 | |||
| M14 | Cytotoxicity assay | Cytotoxicity against human M14 cells expressing NRAS G12C mutant, IC50 = 0.15 μM. | 24471466 | |||
| SK-MEL-28 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human SK-MEL-28 cells harboring BRAF V600E mutant after 68 hrs by MTS assay, IC50 = 0.48 μM. | 24588073 | ||
| insect cell | Function assay | 60 mins | Inhibition of full length human B-Raf V600E mutant expressed in baculovirus infected insect cells assessed as [gamma-33P]incorporation into MEK after 60 mins by scintillation counting, IC50 = 0.031 μM. | 24900315 | ||
| MALME-3M | Function assay | 1 hr | Inhibition of B-Raf V600E mutant-mediated Erk phosphorylation in human MALME-3M cells after 1 hr by fluorescence analysis, IC50 = 0.061 μM. | 24900315 | ||
| A375 | Function assay | 1 hr | Inhibition of B-Raf V600E mutant-mediated Erk phosphorylation in human A375 cells after 1 hr by fluorescence analysis, IC50 = 0.19 μM. | 24900315 | ||
| COLO205 | Cytotoxicity assay | 4 days | Cytotoxicity against human COLO205 cells after 4 days by CellTiter-Glo assay, EC50 = 0.24 μM. | 24900315 | ||
| WM266.4 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human WM266.4 cells after 48 hrs by MTT assay, IC50 = 0.06 μM. | 25267006 | ||
| A375 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A375 cells after 48 hrs by MTT assay, IC50 = 0.19 μM. | 25267006 | ||
| WM1361 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human WM1361 cells after 48 hrs by MTT assay, IC50 = 1.87 μM. | 25267006 | ||
| A375 | Function assay | Inhibition of B-raf V600E mutant in human A375 cells assessed as reduction in ERK1/2 phosphorylation incubated for 90 mins by Western blotting method, IC50 = 0.0331 μM. | 25462267 | |||
| A375 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A375 cells after 72 hrs by CCK8 assay, IC50 = 3.315 μM. | 25462267 | ||
| SK-MEL-2 | Function assay | Inhibition of wild type B-raf in human SK-MEL-2 cells assessed as reduction in ERK1/2 phosphorylation incubated for 90 mins by Western blotting method | 25462267 | |||
| A375 | Function assay | 72 hrs | Inhibition of BRAF V600E mutant in human A375 cells assessed as inhibition of ERK phosphorylation measured after 72 hrs by ELISA assay, IC50 = 0.15 μM. | 25965804 | ||
| A375 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A375 cells after 72 hrs by resazurin assay, IC50 = 0.17 μM. | 25965804 | ||
| A375 | Function assay | 15 mins | Competitive binding affinity to BRAF in human A375 cells after 15 mins in presence of ATP analogue, IC50 = 0.26 μM. | 25965804 | ||
| A375 | Function assay | 15 mins | Competitive binding affinity to ARAF in human A375 cells after 15 mins in presence of ATP analogue, IC50 = 0.95 μM. | 25965804 | ||
| HCT116 | Function assay | Inhibition of KRAS G13D mutant in human HCT116 cells assessed as inhibition of ERK phosphorylation by ELISA, IC50 = 16.6 μM. | 25965804 | |||
| HCT116 | Function assay | 0.34 to 20000 nM | Paradoxical activation of RAS/RAF/MEK signaling pathway in human HCT116 cells expressing wild type BRAF assessed as ERK phosphorylation at 0.34 to 20000 nM | 25965804 | ||
| NZM20 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human NZM20 cells expressing B-Raf V600E mutant isolated from New Zealand metastatic melanoma patient incubated for 68 hrs by SRB assay, IC50 = 0.024 μM. | 26005530 | ||
| NZM07 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human NZM07 cells expressing B-Raf V600E mutant isolated from New Zealand metastatic melanoma patient incubated for 68 hrs by SRB assay, IC50 = 0.036 μM. | 26005530 | ||
| A375 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human A375 cells expressing B-Raf V600E mutant incubated for 68 hrs by MTT assay, IC50 = 0.079 μM. | 26005530 | ||
| COLO205 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human COLO205 cells expressing B-Raf V600E mutant incubated for 68 hrs by MTT assay, IC50 = 0.309 μM. | 26005530 | ||
| SK-MEL-28 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human SK-MEL-28 cells expressing B-Raf V600E mutant incubated for 68 hrs by MTT assay, IC50 = 0.381 μM. | 26005530 | ||
| HT-29 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human HT-29 cells expressing B-Raf V600E mutant incubated for 68 hrs by MTT assay, IC50 = 0.601 μM. | 26005530 | ||
| SK-MEL-1 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human SK-MEL-1 cells expressing B-Raf V600E mutant incubated for 68 hrs by MTT assay, IC50 = 1.499 μM. | 26005530 | ||
| NZM40 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human NZM40 cells expressing wild type B-Raf isolated from New Zealand metastatic melanoma patient incubated for 68 hrs by SRB assay, IC50 = 3.01 μM. | 26005530 | ||
| NZM09 | Antiproliferative assay | 68 hrs | Antiproliferative activity against human NZM09 cells expressing wild type B-Raf isolated from New Zealand metastatic melanoma patient incubated for 68 hrs by SRB assay, IC50 = 8.33 μM. | 26005530 | ||
| A375P | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A375P cells expressing BRAF V600E mutant after 72 hrs by CellTiter-Glo assay, IC50 = 0.37 μM. | 26724730 | ||
| BL21(DE3) | Function assay | Inhibition of N-terminal his-tagged BRAF V600E mutant (448 to 723 residues) (unknown origin) expressed in Escherichia coli BL21(DE3) cells assessed as phosphorylation of biotinylated-MEK by AlphaScreen assay, IC50 = 0.031 μM. | 26852623 | |||
| A375 | Function assay | 1 hr | Inhibition of B-Raf V600E mutant in human A375 cells assessed as ERK phosphorylation preincubated for 1 hr by Western blot method, IC50 = 0.017 μM. | 27085672 | ||
| MIAPaCa2 | Function assay | 1 hr | Inhibition of wild type B-Raf in human MIAPaCa2 cells assessed as reduction in ERK phosphorylation preincubated for 1 hr by Western blot method, EC50 = 2.29 μM. | 27085672 | ||
| COLO205 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human COLO205 cells harboring B-Raf V600E mutant assessed as growth inhibition after 72 hrs by MTT assay, IC50 = 0.044 μM. | 27155899 | ||
| HT-29 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HT-29 cells harboring B-Raf V600E mutant assessed as growth inhibition after 72 hrs by MTT assay, IC50 = 0.156 μM. | 27155899 | ||
| HCT116 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT116 cells harboring wild type B-Raf assessed as growth inhibition after 72 hrs by MTT assay, IC50 = 14.58 μM. | 27155899 | ||
| WM266.4 | Antiproliferative assay | 24 hrs | Antiproliferative activity against human WM266.4 cells assessed as cell viability after 24 hrs by MTT assay, GI50 = 0.21 μM. | 27238841 | ||
| WM266.4 | Antiproliferative assay | 24 hrs | Antiproliferative activity against human WM266.4 cells harboring BRAF V600E mutant assessed as cell growth inhibition after 24 hrs by MTT assay, IC50 = 0.07 μM. | 27634195 | ||
| A375 | Antiproliferative assay | 24 hrs | Antiproliferative activity against human A375 cells assessed as cell growth inhibition after 24 hrs by MTT assay, IC50 = 0.21 μM. | 27634195 | ||
| WM1361 | Antiproliferative assay | 24 hrs | Antiproliferative activity against human WM1361 cells assessed as cell growth inhibition after 24 hrs by MTT assay, IC50 = 1.86 μM. | 27634195 | ||
| SK-MEL-28 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human SK-MEL-28 cells harboring BRAF V600E mutant assessed as concentration required for total growth inhibition measured after 48 hrs resazurin assay, TGI = 2 μM. | 27774137 | ||
| SK-MEL-28 | Growth inhibition assay | 1 hr | Growth inhibition of human SK-MEL-28 cells harboring BRAF V600E mutant preincubated for 1 hr followed by irradiation of 1.13 kW/m2 UV-light for 5 mins measured after 48 hrs resazurin assay | 27774137 | ||
| A375 | Antiproliferative assay | 72 hrs | Antiproliferative activity human A375 cells after 72 hrs by cell titer-glo luminescence assay, IC50 = 0.7 μM. | 28242553 | ||
| COLO205 | Antiproliferative assay | 72 hrs | Antiproliferative activity human COLO205 cells after 72 hrs by cell titer-glo luminescence assay, IC50 = 5.16 μM. | 28242553 | ||
| HepG2 | Antiproliferative assay | 72 hrs | Antiproliferative activity human HepG2 cells after 72 hrs by cell titer-glo luminescence assay, IC50 = 5.48 μM. | 28242553 | ||
| SK-MEL-2 | Antiproliferative assay | 72 hrs | Antiproliferative activity human SK-MEL-2 cells after 72 hrs by cell titer-glo luminescence assay, IC50 = 5.64 μM. | 28242553 | ||
| K562 | Function assay | 1 hr | Stabilization of BRAF in human K562 cells after 1 hr by thermal shift assay, EC50 = 0.79433 μM. | 28280261 | ||
| K562 | Function assay | 1 hr | Stabilization of FECH in human K562 cells after 1 hr by thermal shift assay, EC50 = 5.01187 μM. | 28280261 | ||
| A375M | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A375M cells harboring BRAF V600E mutant after 72 hrs by MTT assay, IC50 = 0.5 μM. | 28458134 | ||
| 1205 Lu | Antiproliferative assay | 72 hrs | Antiproliferative activity against human 1205 Lu cells harboring BRAF V600E mutant after 72 hrs by MTT assay, IC50 = 2 μM. | 28458134 | ||
| A375M | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A375M cells harboring BRAF V600E mutant after 48 hrs by MTT assay, IC50 = 2.05 μM. | 28458134 | ||
| UACC-903 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human UACC-903 cells harboring BRAF V600E mutant after 72 hrs by MTT assay, IC50 = 2.7 μM. | 28458134 | ||
| 1205 Lu | Antiproliferative assay | 48 hrs | Antiproliferative activity against human 1205 Lu cells harboring BRAF V600E mutant after 48 hrs by MTT assay, IC50 = 7.6 μM. | 28458134 | ||
| UACC-903 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human UACC-903 cells harboring BRAF V600E mutant after 48 hrs by MTT assay, IC50 = 12.3 μM. | 28458134 | ||
| CHL-1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CHL-1 cells harboring wild type BRAF after 72 hrs by MTT assay, IC50 = 12.7 μM. | 28458134 | ||
| CHL-1 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human CHL-1 cells harboring wild type BRAF after 48 hrs by MTT assay, IC50 = 20 μM. | 28458134 | ||
| HCT116 | Function assay | Stimulation of BRAF-CRAF dimerization in human HCT116 cells by luciferase complementation assay, EC50 = 0.601 μM. | 28557458 | |||
| Calu6 | Function assay | 3 uM | 2 hrs | Activation of CRAF in human Calu6 cells assessed as increase in MEK phosphorylation at 3 uM after 2 hrs by FRET assay | 28557458 | |
| Sf9 | Function assay | 1 hr | Inhibition of human ZAK (5 to 309 residues) expressed in baculovirus infected Sf9 insect cells using ZAKtide as substrate after 1 hr by mass spectrometry, IC50 = 0.023 μM. | 28586211 | ||
| Sf9 | Function assay | 30 mins | Inhibition of N-terminal GST-tagged recombinant human full-length ZAK expressed in baculovirus infected Sf9 insect cells using MBP as substrate after 30 mins by ADP-Glo assay, IC50 = 0.0314 μM. | 28586211 | ||
| UACC-903 | Cytotoxicity assay | 25 uM | 48 hrs | Cytotoxicity against human UACC-903 cells assessed as cell viability at 25 uM after 48 hrs by MTT assay relative to control, IC50 = 3.6 μM. | 29133035 | |
| CHL-1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CHL-1 cells harboring wild type BRAF after 72 hrs by MTT assay, IC50 = 13.7 μM. | 29133035 | ||
| A375 | Function assay | 96 hrs | Inhibition of BRAF V600E mutant in human A375 cells assessed as reduction in cell proliferation incubated for 96 hrs by MTT assay, IC50 = 0.127 μM. | 29407977 | ||
| A375 | Function assay | Inhibition of BRAF V600E mutant in human A375 cells assessed as reduction in ERK phosphorylation by AlphaScreen assay, IC50 = 0.032 μM. | 29461827 | |||
| SK-MEL-32 | Cytotoxicity assay | Cytotoxicity against human SK-MEL-32 cells, IC50 = 0.31 μM. | 29461827 | |||
| WM266.4 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human WM266.4 cells after 48 hrs by MTT assay, GI50 = 0.21 μM. | 29940463 | ||
| A375 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A375 cells after 48 hrs by MTT assay, GI50 = 0.95 μM. | 29940463 | ||
| HT-29 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HT-29 cells after 48 hrs by MTT assay, GI50 = 1.88 μM. | 29940463 | ||
| WM1361 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human WM1361 cells after 48 hrs by MTT assay, GI50 = 20.8 μM. | 29940463 | ||
| HCT116 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human HCT116 cells after 48 hrs by MTT assay, GI50 = 25.2 μM. | 29940463 | ||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for DAOY cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for A673 cells) | 29435139 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for U-2 OS cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh41 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-MC cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for TC32 cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 489.92 | Formula | C23H18ClF2N3O3S |
Storage (From the date of receipt) | 3 years -20°C(in the dark) powder 1 year -80°C(in the dark) in solvent |
|---|---|---|---|---|---|
| CAS No. | 918504-65-1 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | RG7204, RO5185426,PLX4032 | Smiles | CCCS(=O)(=O)NC1=C(C(=C(C=C1)F)C(=O)C2=CNC3=C2C=C(C=N3)C4=CC=C(C=C4)Cl)F | ||
Solubility
|
In vitro |
DMSO
: 98 mg/mL
(200.03 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Features |
A novel and potent inhibitor of the B-RAFV600E oncoprotein.
|
|---|---|
| Targets/IC50/Ki |
SRMS [1]
(Cell-free assay) 18 nM
ACK1 [1]
(Cell-free assay) 19 nM
B-Raf (V600E) [1]
(Cell-free assay) 31 nM
C-Raf [1]
(Cell-free assay) 48 nM
MAP4K5 (KHS1) [1]
(Cell-free assay) 51 nM
FGR [1]
(Cell-free assay) 63 nM
B-Raf [1]
(Cell-free assay) 100 nM
LCK [1]
(Cell-free assay) 183 nM
BRK [1]
(Cell-free assay) 213 nM
NEK11 [1]
(Cell-free assay) 317 nM
BLK [1]
(Cell-free assay) 547 nM
Lyn B [1]
(Cell-free assay) 599 nM
YES1 [1]
(Cell-free assay) 604 nM
WNK3 [1]
(Cell-free assay) 877 nM
MNK2 [1]
(Cell-free assay) 1.717 μM
FRK (PTK5) [1]
(Cell-free assay) 1.884 μM
CSK [1]
(Cell-free assay) 2.339 μM
Src [1]
(Cell-free assay) 2.389 μM
|
| In vitro |
Vemurafenib (PLX4032) inhibits B-RAFV600E, C-RAF, as well as wildtype B-RAF, with IC50 of 31 nM, 48 nM and 100 nM, respectively. This compound also inhibits several non-RAF kinases, including ACK1, KHS1, and SRMS, with IC50 of 18 nM to 51 nM. [1]
In melanoma cell lines, the inhibitory effect by it depends on B-RAF mutational status, because it potently inhibits those harboring B-RAF V600 mutants, including V600E, V600D, V600K, and V600R, but not wildtype or other mutants. The IC50 values on these cells, including MALME-3M, Colo829, Colo38, A375, SK-MEL28, and A2058, ranges from 20 nM to 1 μM. In these cells, Vemurafenib (0.1 μM to 30 μM) also inhibits the phosphorylation of both MEK1/2 and ERK1/2. [2]
It is highly effective in the treatment of melanoma, for its ability of inhibiting B-RAFV600E. However, this compound displays limited effect in colon cancer patients that also carrying B-RAFV600E oncoprotein. The reason for this is that, in colon cancer cells, B-RAFV600E inhibition by it results in a rapid feedback EGFR activation, which compensates for the PLX4032-inhibited cell proliferation. [3]
|
| Kinase Assay |
RAF kinase activity measurements
|
|
The kinase activities of wild-type RAF and mutants are determined by measuring phosphorylation of biotinylated-BAD protein in the presence of Vemurafenib (PLX4032). For each enzyme (0.01 ng), 20 μL reactions are carried out in 20 mM Hepes (pH 7.0), 10 mM MgCl2, 1 mM DTT, 0.01% (v/v) Tween-20, 50 nM biotin-BAD protein, and 1 mM ATP at room temperature. Reactions are stopped at 5 min with 5 μL of a solution containing 20 mM Hepes (pH 7.0), 200 mM NaCl, 80 mM EDTA, 0.3% (w/v) bovine serum albumin (BSA). The stop solution also includes phospho-BAD (Ser112) antibody, streptavidin-coated donor beads, and protein A acceptor beads. The antibody and beads are pre-incubated in stop solution in the dark at room temperature for 30 min. The final dilution of antibody is 1/2000 and the final concentration of each bead is 10 μg/mL. The assay plates are incubated at room temperature for one hour and then are read on a PerkinElmer AlphaQuest reader. Mutant activities are the average of two different batches of purified protein assayed in duplicate in three different experiments.
|
|
| In vivo | References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
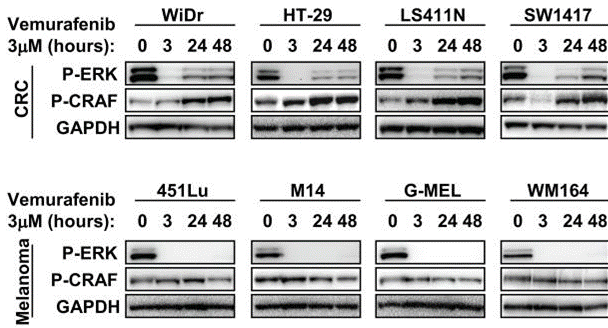
| Western blot | p-ERK / p-CRAF p-MEK(S217/221) / pAKT(T308) / p-AKT(S473) / p-P70 S6K(T389) / p-S6(Ser235-236) / P-4EB-P1 Bax / Bcl2 / Bcl-xl / BIM / Mcl1 |
|
22448344 |
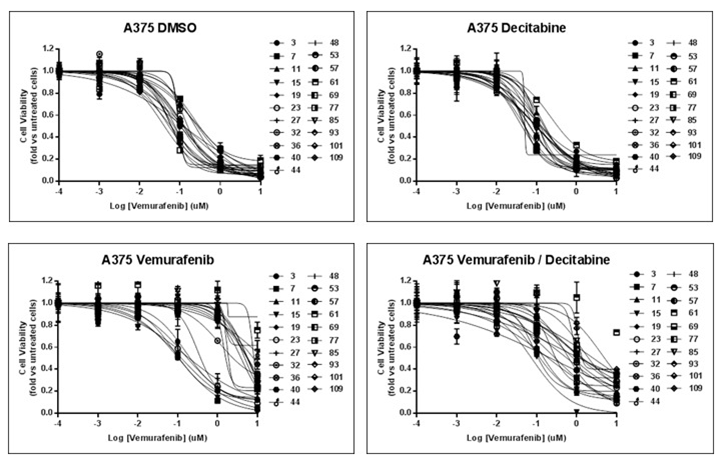
| Growth inhibition assay | Cell viability |
|
29179510 |
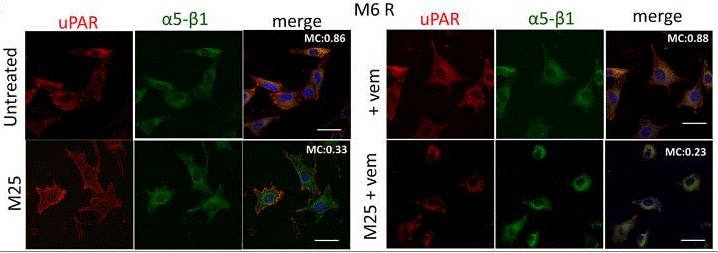
| Immunofluorescence | uPAR / α5-β1 p-Akt(Thr308) |
|
30611716 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT05768178 | Recruiting | Solid Tumor|Haematological Malignancy|Melanoma|Thyroid Cancer Papillary|Ovarian Neoplasms|Colorectal Neoplasms|Laryngeal Neoplasms|Carcinoma Non-Small-Cell Lung|Glioma|Multiple Myeloma|Erdheim-Chester Disease|Thyroid Carcinoma Anaplastic |
Cancer Research UK|University of Manchester|University of Birmingham|Royal Marsden NHS Foundation Trust|Hoffmann-La Roche |
March 1 2023 | Phase 2|Phase 3 |
| NCT05068752 | Recruiting | Pancreas Cancer |
HonorHealth Research Institute|Bayer|Genentech Inc. |
October 28 2021 | Phase 2 |
| NCT03410875 | Active not recruiting | Hairy Cell Leukemia|Leukemia|Leukemia Hairy Cell |
Memorial Sloan Kettering Cancer Center|Dana-Farber Cancer Institute|Yale University |
February 9 2018 | Phase 2 |
| NCT03013491 | Completed | Solid Tumor|Lymphoma |
CytomX Therapeutics |
January 2017 | Phase 1|Phase 2 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
How about its half-life?
Answer:
It was reported that this compound has a half-life of 57 hours.
Question 2:
When prepared in 4% DMSO/30% PEG 300/5% Tween 80/ddH2O solutions, it forms a pellet down the tube?
Answer:
When preparing this kind of vehicle, please dissolve it in DMSO clearly first. If it dissolves not readily, please sonicate and warm in the water bath at about 45 degree. Then add PEG and Tween. After they mixed homogeneously, then dilute with water.






































