
- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at [email protected] to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly FLAG Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
Geldanamycin
Synonyms: NSC 122750
Geldanamycin is a natural existing HSP90 inhibitor with Kd of 1.2 μM, specifically disrupts glucocorticoid receptor (GR)/HSP association. Geldanamycin attenuates virus infection-induced ALI (acute lung injury)/ARDS (acute respiratory distress syndrome) by reducing the host's inflammatory responses.
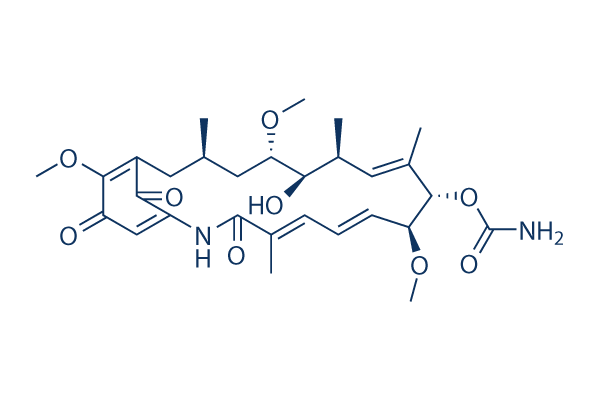
Geldanamycin Chemical Structure
CAS: 30562-34-6
Selleck's Geldanamycin has been cited by 32 publications
Purity & Quality Control
Batch:
Purity:
99.96%
99.96
Geldanamycin Related Products
| Related Compound Libraries | Kinase Inhibitor Library Angiogenesis Related compound Library TGF-beta/Smad compound library Cytoskeletal Signaling Pathway Compound Library Anti-Cardiovascular Disease Compound Library | Click to Expand |
|---|
Choose Selective Antineoplastic and Immunosuppressive Antibiotics Inhibitors
Cell Data
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| A2780 cells | Proliferation assay | Compound was evaluated for antiproliferative activity against human ovarian carcinoma cell line A2780, IC50=3.4 μM | 11514145 | |||
| SW620 cell | Growth inhibition assay | Inhibitory concentration against human colorectal carcinoma SW620 cell lines, IC50=6.2 nM | 15658879 | |||
| MCF-7 cell | Growth inhibition assay | Inhibitory concentration against human breast cancer MCF-7 cell lines, IC50=6.5 nM | 15658879 | |||
| K562 cell | Growth inhibition assay | Inhibitory concentration against human leukemia K562 cell lines, IC50=22.1 nM | 15658879 | |||
| HT-29 cell | Growth inhibition assay | Inhibitory concentration against human colorectal carcinoma HT-29 cell lines, IC50=24.5 nM | 15658879 | |||
| SK-BR-3 | Function assay | 50 mg/kg | Inhibition of oncogene product p185 erbB-2 from human breast cancer cells(SK-BR-3 cells) at 50 mg/kg dose, IC50=0.07μM | 7562911 | ||
| SK-BR-3 | Function assay | In vitro inhibition of p185 erbB-2 oncoprotein in human breast cancer SK-BR-3 cells, IC50=0.07μM | 7562912 | |||
| MCF-7 | Function assay | Inhibition of steady-state expression of HER2 in MCF-7 breast cancer cells, IC50=0.05μM | 10340605 | |||
| MCF-7 | Function assay | Inhibition of steady-state expression of ER in MCF-7 breast cancer cells, IC50=0.06μM | 10340605 | |||
| MCF-7 | Function assay | Inhibition of steady-state expression of Raf-1 in MCF-7 breast cancer cells, IC50=0.2μM | 10340605 | |||
| SKBR-3 | Function assay | Effective concentration for Her-2 degradation in SKBR-3 cells, Concentration=5μM | 15713410 | |||
| HEK293 | Function assay | Inhibition of HSP90 expressed in HEK293 cells assessed as effect on Akt1:p27 interaction complexes by EYFP and/or YFP Venus fragment based reporter gene assay | 16680159 | |||
| LS174T | Cytotoxicity assay | Cytotoxicity against human LS174T cells by MTS assay, IC50=0.45μM | 17034135 | |||
| rat neuronal cells | Cytotoxicity assay | 10 uM | 24 hrs | Cytotoxicity against rat neuronal cells at 10 uM after 24 hrs | 17276679 | |
| P19 | Cytotoxicity assay | 18 hrs | Cytotoxicity against mouse P19 cells after 18 hrs, IC50=0.1μM | 17442565 | ||
| MCF7 | Growth inhibition assay | Growth inhibition of human MCF7 cells after days by SRB assay, GI50=35.6μM | 17869098 | |||
| HCT116 | Growth inhibition assay | Growth inhibition of human HCT116 cells, GI50=0.021μM | 18243703 | |||
| DLD1 | Growth inhibition assay | Growth inhibition of human DLD1 cells, GI50=0.037μM | 18243703 | |||
| KPL4 | Growth inhibition assay | Growth inhibition of human KPL4 cells, GI50=0.059μM | 18243703 | |||
| A549 | Growth inhibition assay | Growth inhibition of human A549 cells, GI50=0.064μM | 18243703 | |||
| KPL4 | Function assay | 40 hrs | Inhibition of Hsp90 in human KPL4 cells assessed as depletion of ErbB2 level after 40 hrs, MEC=0.11μM | 18243703 | ||
| KPL4 | Function assay | 40 hrs | Inhibition of Hsp90 in human KPL4 cells assessed as depletion of Raf-1 level after 40 hrs, MEC=0.33μM | 18243703 | ||
| Jurkat | Function assay | 1 uM | Inhibition of to HSP90 in FADD deficient human Jurkat cells assessed as RIP protein degradation at 1 uM by Western blot | 18408713 | ||
| HEK293T | Function assay | Inhibition of TNF-alpha-induced NF-kappaB activation expressed in HEK293T cells by luciferase reporter gene assay | 18408713 | |||
| Jurkat | Function assay | Inhibition of TNF-alpha-induced NF-kappaB activation expressed in FADD deficient human Jurkat cells by luciferase reporter gene assay | 18408713 | |||
| SKBR3 | Function assay | 24 hrs | Inhibition of Hsp90-mediated HER2 degradation in human SKBR3 cells after 24 hrs by Western blot | 18816111 | ||
| SKBR3 | Function assay | 24 hrs | Inhibition of Hsp90-mediated Raf degradation in human SKBR3 cells after 24 hrs by Western blot | 18816111 | ||
| HuH7 | Antiviral assay | 3 days | Antiviral activity against Hepatitis C virus genotype 1b Con1 infected in human HuH7 cells assessed as GAPDH RNA or 18S rRNA level after 3 days by qRT-PCR analysis, EC50=0.00025μM | 18936191 | ||
| HuH7 | Antiviral assay | 3 days | Antiviral activity against Hepatitis C virus genotype 1b Con1 infected in human HuH7 cells assessed as GAPDH RNA or 18S rRNA level after 3 days selected with 40 nM HCV-796 and 800 nM boceprevir by qRT-PCR analysis, EC50=0.0012μM | 18936191 | ||
| SH-SY5Y | Neuroprotection assay | Neuroprotection against beta-amyloid peptide 1-42-induced toxicity in human SH-SY5Y cells assessed as lactate dehydrogenase release, EC50=0.04269μM | 19138859 | |||
| SH-SY5Y | Neuroprotection assay | 10 nM | Neuroprotection against beta-amyloid peptide 1-42-induced toxicity in retinoic acid-differentiated human SH-SY5Y cells assessed as lactate dehydrogenase release at 10 nM | 19138859 | ||
| SKBR3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SKBR3 cells after 72 hrs by celltiter-glo assay, IC50=0.041μM | 19405528 | ||
| MCF7 | Cytotoxicity assay | Cytotoxicity against human MCF7 cells by SRB assay, IC50=9.6μM | 19560353 | |||
| SK-BR-3 | Antiproliferative assay | Antiproliferative activity against human SK-BR-3 cells, IC50=0.0158μM | 19896848 | |||
| MCF7 | Antiproliferative assay | Antiproliferative activity against human MCF7 cells, IC50=0.0161μM | 19896848 | |||
| A431 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A431 cells after 72 hrs, IC50=0.2μM | 20655237 | ||
| PP30 | Function assay | 100 uM | 3 hrs | Inhibition of human HSP90alpha expressed in yeast PP30 cells co-expressing HSF-lacZ reporter assessed as induction of heat shock response at 100 uM after 3 hrs | 21129982 | |
| MCF7 | Function assay | 0.5 uM | Inhibition of Hsp90 in human MCF7 cells assessed as Akt degradation at 0.5 uM by Western blot analysis | 21273068 | ||
| MCF7 | Function assay | 0.5 uM | Inhibition of Hsp90 in human MCF7 cells assessed as Raf degradation at 0.5 uM by Western blot analysis | 21273068 | ||
| MCF7 | Function assay | 0.5 uM | Inhibition of Hsp90 in human MCF7 cells assessed as Hsp90 degradation at 0.5 uM by Western blot analysis | 21273068 | ||
| MCF7 | Function assay | 0.5 uM | Inhibition of Hsp90 in human MCF7 cells assessed as Her2 degradation at 0.5 uM by Western blot analysis | 21273068 | ||
| HCT116 | Antiproliferative assay | 4 days | Antiproliferative activity against human HCT116 cells measured on day 4 by Cell titer-glo assay, EC50=0.03μM | 21420297 | ||
| HCT116 | Antiproliferative assay | Antiproliferative activity against human HCT116 cells by luminescence assay, EC50=0.03μM | 21605975 | |||
| U87 | Antiproliferative assay | Antiproliferative activity against human U87 cells by luminescence assay, EC50=0.089μM | 21605975 | |||
| HCT116 | Antiproliferative assay | Antiproliferative activity against human HCT116 cells, IC50=0.03μM | 21715165 | |||
| NCI-H1975 | Antiproliferative assay | Antiproliferative activity against human NCI-H1975 cells, IC50=0.036μM | 21715165 | |||
| U87MG | Antiproliferative assay | Antiproliferative activity against human U87MG cells, IC50=0.089μM | 21715165 | |||
| MCF7 | Function assay | 500 nM | 24 hrs | Inhibition of Hsp90 in human MCF7 cells assessed as induction of Her2 degradation at 500 nM after 24 hrs by Western blot method | 21861487 | |
| MCF7 | Function assay | 500 nM | 24 hrs | Inhibition of Hsp90 in human MCF7 cells assessed as induction of Raf degradation at 500 nM after 24 hrs by Western blot method | 21861487 | |
| MCF7 | Function assay | 500 nM | 24 hrs | Inhibition of Hsp90 in human MCF7 cells assessed as induction of Akt degradation at 500 nM after 24 hrs by Western blot method | 21861487 | |
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MCF7 cells at 72 hrs by MTS assay, IC50=0.053μM | 22162786 | ||
| HepG2 | Cytotoxicity assay | Cytotoxicity against human HepG2 cells, IC50=0.37μM | 22849774 | |||
| SKBR3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against estrogen receptor deficient human SKBR3 cells after 72 hrs, IC50=0.0085μM | 23648180 | ||
| MCF7 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MCF7 cells expressing estrogen receptor after 72 hrs, IC50=0.0098μM | 23648180 | ||
| HepG2 | Cytotoxicity assay | Cytotoxicity against human HepG2 cells by MTT assay, IC50=0.3μM | 23656556 | |||
| SKBR3 | Function assay | 0.5 uM | 18 hrs | Inhibition of HSP90 in human SKBR3 cells assessed as aggregation of intracellular HER2 at 0.5 uM after 18 hrs by DAPI staining-based immunofluorescence microscopic analysis | 23859777 | |
| A549 | Cytotoxicity assay | 2 days | Cytotoxicity against human A549 cells after 2 days by AlamarBlue assay, IC50=0.15μM | 23947794 | ||
| CL1-5 | Function assay | 2 uM | 16 hrs | Inhibition of HSP90 in human CL1-5 cells assessed as reduction of total Akt at 2 uM after 16 hrs by Western blotting analysis | 24428777 | |
| L6 | Cytotoxicity assay | 72 hrs | Cytotoxicity against rat L6 cells after 72 hrs by Alamar Blue assay, IC50=6μM | 24580531 | ||
| Sf9 | Function assay | 16 hrs | Displacement of GM-BODIPY from human full length HSP90 alpha expressed in baculovirus-infected Sf9 cells after 16 hrs by fluorescence polarization assay, IC50=0.074μM | 24751441 | ||
| MDA-kb2 | Function assay | 18 hrs | Inhibition of HSP90 in human MDA-kb2 cells assessed as reduction in glucocorticoid receptor-dependent luciferase expression after 18 hrs by firefly luciferase reporter gene assay, IC50<0.01μM | 24984936 | ||
| MCF7 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MCF7 cells assessed as growth inhibition after 72 hrs by MTS/PMS assay, IC50=0.04μM | 25075762 | ||
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-231 cells after 72 hrs by MTT assay, IC50=0.05μM | 25105924 | ||
| LNCAP | Cytotoxicity assay | 72 hrs | Cytotoxicity against human LNCAP cells after 72 hrs by MTT assay, IC50=0.43μM | 25105924 | ||
| MDA-MB 231 | Function assay | 0.5 and 5 uM | Inhibition of HSP90 in human MDA-MB 231 cells assessed increase in HSP70 protein levels at 0.5 and 5 uM by Western blot method | 25105924 | ||
| HUVEC | Cytotoxicity assay | 72 hrs | Cytotoxicity against HUVEC cells after 72 hrs by MTT assay, IC50=0.019μM | 25277067 | ||
| A431 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A431 cells after 72 hrs by MTT assay, IC50=0.04μM | 25277067 | ||
| BGC823 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human BGC823 cells after 72 hrs by MTT assay, IC50=0.04μM | 25277067 | ||
| HepG2 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HepG2 cells after 72 hrs by MTT assay, IC50=0.04μM | 25277067 | ||
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-231 cells after 72 hrs by MTT assay, IC50=0.05μM | 25277067 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells after 72 hrs by MTT assay, IC50=0.097μM | 25277067 | ||
| HL7702 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HL7702 cells after 72 hrs by MTT assay, IC50=0.141μM | 25277067 | ||
| SW480 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SW480 cells after 72 hrs by MTT assay, IC50=0.31μM | 25277067 | ||
| HeLa | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HeLa cells after 72 hrs by MTT assay, IC50=0.798μM | 25277067 | ||
| MDA-MB 231 | Function assay | 0.5 uM | 24 hrs | Inhibition Hsp90 in human MDA-MB 231 cells assessed as increase in HSP70 protein level at 0.5 uM incubated for 24 hrs by Western blot method | 25277067 | |
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MCF7 cells assessed as growth inhibition after 72 hrs by MTS/PMS assay, IC50=0.0427μM | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as pAkt degradation at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as Her2 degradation at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as Cdk6 degradation at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as Raf degradation at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Change in Cdc37 expression in human MCF7 cells at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Change in p23 expression in human MCF7 cells at 5 times IC50 after 24 hrs by Western blot analysis | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as induction of heat shock response-mediated HSP90 production at 5 times IC50 after 24 hrs by Western blot analysis relative to vehicle-treated control | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as induction of heat shock response-mediated HSP70 production at 5 times IC50 after 24 hrs by Western blot analysis relative to vehicle-treated control | 25756299 | ||
| MCF7 | Function assay | 24 hrs | Inhibition of HSP90 in human MCF7 cells assessed as induction of heat shock response-mediated HSP27 production at 5 times IC50 after 24 hrs by Western blot analysis relative to vehicle-treated control | 25756299 | ||
| PC12 | Function assay | 10 uM | 3 to 16 hrs | Inhibition of D-aspartate biosynthesis in rat PC12 cells assessed as reduction of intracellular D-aspartate content at 10 uM measured at 3 to 16 hrs by O-phthalaldehyde/N-acetyl-L-cysteine derivatization technique-based HPLC analysis relative to vehicle-t | 26642769 | |
| PC12 | Function assay | 10 uM | 6 to 16 hrs | Inhibition of D-aspartate biosynthesis in rat PC12 cells assessed as reduction of total D-aspartate content at 10 uM measured at 6 to 16 hrs by O-phthalaldehyde/N-acetyl-L-cysteine derivatization technique-based HPLC analysis relative to vehicle-treated c | 26642769 | |
| PC12 | Function assay | 24 hrs | Inhibition of D-aspartate biosynthesis in rat PC12 cells assessed as reduction of intracellular D-aspartate level up to 10 uM after 24 hrs by O-phthalaldehyde/N-acetyl-L-cysteine derivatization technique-based HPLC analysis | 26642769 | ||
| PC12 | Function assay | up to 10 uM | 24 hrs | Inhibition of D-aspartate biosynthesis in rat PC12 cells assessed as reduction of D-aspartate level in cell culture medium up to 10 uM after 24 hrs by O-phthalaldehyde/N-acetyl-L-cysteine derivatization technique-based HPLC analysis | 26642769 | |
| A549 | Function assay | 12 to 24 hrs | Inhibition of Hsp90 in human A549 cells assessed as decrease in EGFR levels at five times IC50 value after 12 to 24 hrs by Western blot analysis | 26745854 | ||
| A549 | Function assay | 12 to 24 hrs | Inhibition of Hsp90 in human A549 cells assessed as decrease in Her2 levels at five times IC50 value after 12 to 24 hrs by Western blot analysis | 26745854 | ||
| A549 | Function assay | 12 to 24 hrs | Inhibition of Hsp90 in human A549 cells assessed as decrease in C-Raf levels at five times IC50 value after 12 to 24 hrs by Western blot analysis | 26745854 | ||
| SKBR3 | Function assay | Inhibition of Hsp90 in human SKBR3 cells, IC50=0.07μM | 26844689 | |||
| BA/F3 | Function assay | 48 hrs | Inhibition of imatinib-resistant BCR-ABL T315I mutant (unknown origin) expressed in mouse BA/F3 cells assessed as decrease in cell viability after 48 hrs by trypan blue exclusion assay, IC50=1μM | 26844689 | ||
| BA/F3 | Function assay | 48 hrs | Inhibition of imatinib-resistant BCR-ABL E255K mutant (unknown origin) expressed in mouse BA/F3 cells assessed as decrease in cell viability after 48 hrs by trypan blue exclusion assay, IC50=1μM | 26844689 | ||
| BA/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL (unknown origin) expressed in mouse BA/F3 cells assessed as decrease in cell viability after 48 hrs by trypan blue exclusion assay, IC50=5μM | 26844689 | ||
| MDA-MB-231 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MDA-MB-231 cells after 72 hrs by MTS/PMS assay, Activity=0.06μM | 27003516 | ||
| MDA-MB-231 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MDA-MB-231 cells after 72 hrs by SRB assay, IC50=0.03μM | 27266997 | ||
| HeLa | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HeLa cells after 72 hrs by SRB assay, IC50=0.06μM | 27266997 | ||
| SNU638 | Anticancer assay | 72 hrs | Anticancer activity against human SNU638 cells measured after 72 hrs by SRB assay, IC50=0.03μM | 27283788 | ||
| Caki1 | Anticancer assay | 72 hrs | Anticancer activity against human Caki1 cells measured after 72 hrs by SRB assay, IC50=0.056μM | 27283788 | ||
| A549 | Anticancer assay | 72 hrs | Anticancer activity against human A549 cells measured after 72 hrs by SRB assay, IC50=0.109μM | 27283788 | ||
| MDA-MB-231 | Anticancer assay | 72 hrs | Anticancer activity against human MDA-MB-231 cells measured after 72 hrs by SRB assay, IC50=0.11μM | 27283788 | ||
| HCT116 | Anticancer assay | 72 hrs | Anticancer activity against human HCT116 cells measured after 72 hrs by SRB assay, IC50=0.15μM | 27283788 | ||
| SKBR3 | Antiproliferative assay | 1 to 3 days | Antiproliferative activity against Her2-overexpressing human SKBR3 cells after 1 to 3 days by MTS assay, GI50=0.43μM | 27783977 | ||
| NCI-H1975 | Antiproliferative assay | 1 to 3 days | Antiproliferative activity against gefitinib-resistant human NCI-H1975 cells after 1 to 3 days by MTS assay, GI50=0.56μM | 27783977 | ||
| RPMI8226 | Growth inhibition assay | 72 hrs | Growth inhibition of human RPMI8226 cells after 72 hrs by MTS/PMS assay, GI50=0.003μM | 29057043 | ||
| MDA-MB-231 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MDA-MB-231 cells after 72 hrs by SRB assay, IC50=0.06μM | 29172085 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by SRB assay, IC50=0.1μM | 29172085 | ||
| NCI-H1975 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human NCI-H1975 cells after 72 hrs by Cell Titer 96 Aqueous One Solution cell proliferation assay, GI50=0.56μM | 29202402 | ||
| A549 | Function assay | 5 to 10 uM | 24 hrs | Inhibition of HSP90alpha in human A549 cells assessed as upregulation of HSP90alpha expression at 5 to 10 uM after 24 hrs by Western blot analysis | 29486954 | |
| A549 | Function assay | 5 to 10 uM | 24 hrs | Inhibition of HSP90beta in human A549 cells assessed as upregulation of HSP90beta expression at 5 to 10 uM after 24 hrs by Western blot analysis | 29486954 | |
| SKBR3 | Function assay | 24 hrs | Inhibition of HSP90 at N-terminus in human SKBR3 cells assessed as induction of Raf1 protein degradation incubated for 24 hrs by Western blot analysis | 31591016 | ||
| SKBR3 | Function assay | 24 hrs | Inhibition of HSP90 at N-terminus in human SKBR3 cells assessed as induction of CDK4 protein degradation incubated for 24 hrs by Western blot analysis | 31591016 | ||
| SKBR3 | Function assay | 24 hrs | Inhibition of HSP90 at N-terminus in human SKBR3 cells assessed as induction of Her2 protein degradation incubated for 24 hrs by Western blot analysis | 31591016 | ||
| SKBR3 | Function assay | 24 hrs | Inhibition of HSP90 at N-terminus in human SKBR3 cells assessed as induction of AKT protein degradation incubated for 24 hrs by Western blot analysis | 31591016 | ||
| Click to View More Cell Line Experimental Data | ||||||
Biological Activity
| Description | Geldanamycin is a natural existing HSP90 inhibitor with Kd of 1.2 μM, specifically disrupts glucocorticoid receptor (GR)/HSP association. Geldanamycin attenuates virus infection-induced ALI (acute lung injury)/ARDS (acute respiratory distress syndrome) by reducing the host's inflammatory responses. | ||||||
|---|---|---|---|---|---|---|---|
| Targets |
|
| In vitro | ||||
| In vitro | Geldanamycin binds in the ATP-binding site in the N-terminus domain of Hsp90s (residues 1-220). Geldanamycin inhibits the ATPase activity of Hsp90 in a dose-dependent manner. [1] Geldanamycin causes a dose-dependent G2 arrest and reversible inhibiton o f entry into the S phase in A2780 human ovarian cell line. This inhibition is accompanied by p53 increase and finally demonstrated to be p53 dependent. [2] Geldanamycin causes polyubiquitination and proteasomal degradation of the p185 receptor protein-tyrosin kinase and shows a IC50 with 70 nM. [3, 4] Geldanamycin is a typical anti-tumor reagent, shows a mean GI50 with 0.18 μM against the panel of 60 human tumor cell lines. [5] | |||
|---|---|---|---|---|
| Kinase Assay | Isothermal Titration Calorimetry (ITC) of Nucelotide Binding | |||
| The titration experiments are performed using the MSC system. In each experiment, 16 aliquots of 15 μL of geldanamycin (300 μM in 1% DMSO) are injected into 1.3 mL of protein (31 μM in 20 mMTris-HCl, pH 7.5, 1 mMEDTA) at 25 °C, and the resulting data are fit after subtracting the heats of dilution. Heats of dilution are determined in separate experiments from addition of geldanamycin into buffer and buffer into protein. No evidence for binding of DMSO in the nucleotide binding site is observed. Titration data are fit using a nonlinear least-squares curve-fitting algorithm with three floating variables: stoichiometry, binding constant (Kb) 1/Kd), and change of enthalpy of interaction (ΔH°). Dissociation constants estimated for geldanamycin binding to intact yeast Hsp90 is 1.22 μM, and for binding to Hsp90 N-terminal domain is 0.78 μM. No meaningful heat is observed with binding to the C-terminal fragment. | ||||
| Cell Research | Cell lines | A2780 human ovarian cell line | ||
| Concentrations | 0.001-10 μM | |||
| Incubation Time | 3 hours | |||
| Method | Exponentially growing cells are treated with Geldanamycin and at various times DNA synthesis is assessed by incorporation of bromodeoxyuridine (BrdUrd) and flow cytometric analysis. No marked difference in total cell number is noted during this time course for treated and untreated cultures. BrdUrd (10 μM) is incorporated over a 4-h incubation period at 37 °C and cells are harvested and fixed in 70% ethanol. After denaturation of the DNA with 2 N HC1, cells are incubated with an anti-BrdUrd mouse monoclonal antibody followed by a fluorescein isothiocyanate (FITC)-linked goat anti-mouse IgG. Cells are stained for 30 minutes at room temperature with propidium iodide and analysed by flow cytometry using a Coulter EPICS Profile Analyzer. |
|||
| In Vivo | ||
| In vivo | Geldanamycin (50 mg//kg) shows 30% inhibition on pl85-associated phosphotyrosine levels in FRE/erbB-2 mice. [6] | |
|---|---|---|
| Animal Research | Animal Models | FRE/erbB-2 tumors in nu/nu mice |
| Dosages | 50 mg/kg | |
| Administration | Administered via i.p. | |
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT00003969 | Completed | Unspecified Adult Solid Tumor Protocol Specific |
Cancer Research UK|National Cancer Institute (NCI) |
August 1998 | Phase 1 |
Chemical Information & Solubility
| Molecular Weight | 560.64 | Formula | C29H40N2O9 |
| CAS No. | 30562-34-6 | SDF | Download Geldanamycin SDF |
| Smiles | CC1CC(C(C(C=C(C(C(C=CC=C(C(=O)NC2=CC(=O)C(=C(C1)C2=O)OC)C)OC)OC(=O)N)C)C)O)OC | ||
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 100 mg/mL ( (178.36 mM); Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Water : Insoluble Ethanol : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field
Tags: buy Geldanamycin | Geldanamycin supplier | purchase Geldanamycin | Geldanamycin cost | Geldanamycin manufacturer | order Geldanamycin | Geldanamycin distributor







































