- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at [email protected] to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly FLAG Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
PF-573228
PF-573228 is an ATP-competitive inhibitor of FAK with IC50 of 4 nM in a cell-free assay, ~50- to 250-fold selective for FAK than Pyk2, CDK1/7 and GSK-3β. PF-573228 induces apoptosis.
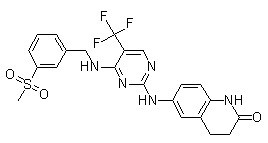
PF-573228 Chemical Structure
CAS: 869288-64-2
Selleck's PF-573228 has been cited by 82 publications
Purity & Quality Control
Batch:
Purity:
99.92%
99.92
PF-573228 Related Products
| Related Compound Libraries | Tyrosine Kinase Inhibitor Library PI3K/Akt Inhibitor Library Angiogenesis Related compound Library HIF-1 Signaling Pathway Compound Library FDA-approved Anticancer Drug Library | Click to Expand |
|---|
Signaling Pathway
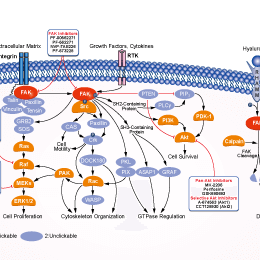
Choose Selective FAK Inhibitors
Cell Data
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| A431 | Kinase assay | ~10 μM | DMSO | inhibits FAK phosphorylation with IC50 of 11 nM | 17395594 | |
| REF52 | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of ~100 nM | 17395594 | |
| PC3 | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 100 nM | 17395594 | |
| SKOV-3 | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 50 nM | 17395594 | |
| L3.6p1 | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 300 nM | 17395594 | |
| F-G | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 30 nM | 17395594 | |
| MDCK | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 500 nM | 17395594 | |
| PC3 | Growth inhibitory assay | 10 μM | DMSO | significantly inhibits cell growth. | 17395594 | |
| REF52 | Growth inhibitory assay | 10 μM | DMSO | significantly inhibits cell growth. | 17395594 | |
| MDCK | Apoptosis assay | 10 μM | DMSO | induces apoptosis | 17395594 | |
| REF52 | Apoptosis assay | 10 μM | DMSO | induces apoptosis | 17395594 | |
| REF52 | Function assay | 10 μM | DMSO | blocks serum and FN-stimulated migration | 17395594 | |
| platelet | Function assay | 1 μM | DMSO | inhibits platelet aggregation and spreading | 19716803 | |
| platelet | Function assay | 1 μM | DMSO | leads to inhibition of PAK and AKT | 19716803 | |
| platelet | Function assay | 1 μM | DMSO | blocks calcium mobilization and dense granule secretion | 19716803 | |
| 4T1 | Function assay | DMSO | abolishes the interaction between β3 integrin and TβR-II | 19740433 | ||
| MCF7 | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 430 nM | 20354780 | |
| TamR | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 50 nM | 20354780 | |
| FasR | Kinase assay | ~10 μM | DMSO | inhibits the phosphorylation of FAK Tyr397 with IC50 of 130 nM | 20354780 | |
| TamR | Function assay | 1 μM | DMSO | inhibits cell migration | 20354780 | |
| FasR | Function assay | 1 μM | DMSO | inhibits cell migration | 20354780 | |
| endothelial cell | Kinase assay | 40 nM | DMSO | inhibits H2O2-induced phosphorylation of FAK | 21212402 | |
| endothelial cell | Function assay | 40 nM | DMSO | inhibits H2O2-induced stress fiber formation | 21212402 | |
| endothelial cell | Apoptosis assay | 40 nM | DMSO | inhibits apoptosis | 21212402 | |
| GH3 | Function assay | 3 μM | DMSO | increases IK(Ca) amplitude | 21925512 | |
| GH3 | Function assay | 3 μM | DMSO | enhances BKCa-channel activity | 21925512 | |
| HUVEC | cytotoxicity assay | ~10 μM | DMSO | impairs endothelial cell viability | 22075057 | |
| HUVEC | Kinase assay | 5 μM | DMSO | inhibits FAK kinase activity | 22075057 | |
| HUVEC | Function assay | 5 μM | DMSO | induces cell cycle arrest | 22075057 | |
| HUVEC | Apoptosis assay | 5 μM | DMSO | induces apoptosis | 22075057 | |
| HUVEC | Function assay | 5 μM | DMSO | impedes endothelial cell migration and alters the cellular actin cytoskeleton | 22075057 | |
| HUVEC | Function assay | 5 μM | DMSO | blocks HUVEC sprouting on collagen I gels | 22075057 | |
| human peripheral blood T cells | Kinase assay | ~10 μM | DMSO | inhibits site-specific phosphorylation of FAK | 23928188 | |
| human peripheral blood T cells | Function assay | ~10 μM | DMSO | impairs TCR-induced T cell morphological changes and alters activity of RhoA | 23928188 | |
| human peripheral blood T cells | Function assay | ~10 μM | DMSO | inhibits phosphorylation of ZAP-70 and LAT | 23928188 | |
| human peripheral blood T cells | Function assay | ~10 μM | DMSO | impairs Antigen-dependent T cell conjugation | 23928188 | |
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for U-2 OS cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Saos-2 cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| Click to View More Cell Line Experimental Data | ||||||
Biological Activity
| Description | PF-573228 is an ATP-competitive inhibitor of FAK with IC50 of 4 nM in a cell-free assay, ~50- to 250-fold selective for FAK than Pyk2, CDK1/7 and GSK-3β. PF-573228 induces apoptosis. | ||
|---|---|---|---|
| Targets |
|
| In vitro | ||||
| In vitro | PF 573228 blocks the phosphorylation of FAK Tyr397 in REF52 cells, PC3 cells, SKOV-3 cells, L3.6p1 and F-G, MDCK cells with IC50 of 30-500 nM. However, PF 573228 (1 μM) with 80% inhibition of FAK phosphorylation fails to inhibit cell growth or apoptosis. Similar treatment of cells with PF-228 resulted in inhibition of serum or FN-directed migration and decreased focal adhesion turnover. [1] | |||
|---|---|---|---|---|
| Kinase Assay | Affinity determination | |||
| Purified activated FAK kinase domain (amino acids 410–689) is reacted with 50 μM ATP, and 10 μg/well of a random peptide polymer of Glu and Tyr (molar ratio of 4:1), poly(Glu/Tyr) in kinase buffer (50 mM HEPES, pH 7.5, 125 mM NaCl, 48 mM MgCl2) for 15 min. Phosphorylation of poly(Glu/Tyr) is challenged with serially diluted compounds at 1/2-Log concentrations starting at a top concentration of 1 μM. Each concentration is run in triplicate. Phosphorylation of poly(Glu/Tyr) is detected with a general anti-phospho-tyrosine (PY20) antibody, followed by horseradish peroxidase-conjugated goat anti-mouse IgG antibody. The standard horseradish peroxidase substrate 3, 3 | ||||
| Cell Research | Cell lines | REF52 or PC3 cells | ||
| Concentrations | ~10 μM | |||
| Incubation Time | 3 days | |||
| Method | Growth assays are performed by seeding 1 × 104 REF52 or PC3 cells/well of a 24-well plate in triplicate 24 h prior to daily treatment with the indicated concentrations of each inhibitor for 3 days. Subsequently, the cells are harvested and counted. |
|||
| Experimental Result Images | Methods | Biomarkers | Images | PMID |
| Western blot | cyclin B1 p-FAK / FAK Lamin A / Lamin C |

|
30761269 | |
| Immunofluorescence | FAK / F-actin Emerin |
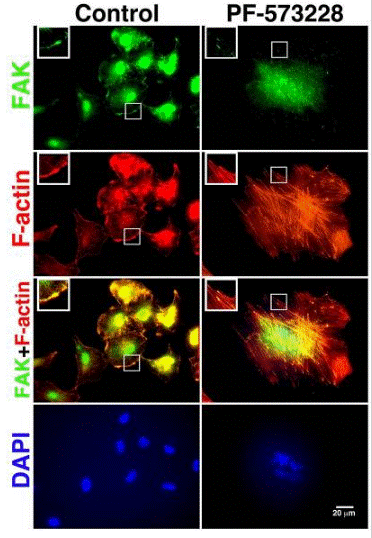
|
30761269 | |
| Growth inhibition assay | Cell viability |
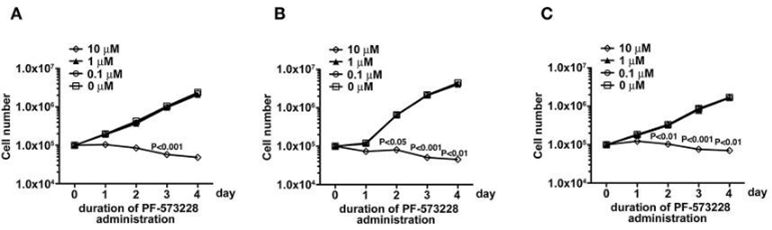
|
30761269 | |
| In Vivo | ||
| In vivo | Inhibition of FAK by PF-573,228 in Ctrl-MT mice leads to a significant suppression of mammary tumorigenesis as well as lung metastasis. In contrast, treatment of MFCKO-MT mice with PF-573,228 did not affect the initiation of mammary tumors in these mice, as would be expected due to the absence of FAK in mammary epithelial cells of these mice [2]. | |
|---|---|---|
| Animal Research | Animal Models | Ctrl-MT and MFCKO-MT mice |
| Dosages | 5 mg/kg | |
| Administration | oral administration | |
Chemical Information & Solubility
| Molecular Weight | 491.49 | Formula | C22H20F3N5O3S |
| CAS No. | 869288-64-2 | SDF | Download PF-573228 SDF |
| Smiles | CS(=O)(=O)C1=CC=CC(=C1)CNC2=NC(=NC=C2C(F)(F)F)NC3=CC4=C(C=C3)NC(=O)CC4 | ||
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 26 mg/mL ( (52.9 mM); Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Water : Insoluble Ethanol : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field
Frequently Asked Questions
Question 1:
Would you please let me know the detail of how to dissolve PF-573228 (Catalog No.S2013) for in vivo study (oral administration)?
Answer:
PF-573228 in 30% PEG400+0.5% Tween80+ 5% Propylene glycol at 30mg/ml is a suspension. If you will use the compound for oral gavage, this suspension is fine for it.
Tags: buy PF-573228 | PF-573228 supplier | purchase PF-573228 | PF-573228 cost | PF-573228 manufacturer | order PF-573228 | PF-573228 distributor