
- Inhibitors
- By product type
- Natural Products
- Inducing Agents
- Peptides
- Antibiotics
- Antibody-drug Conjugates(ADC)
- PROTAC
- Hydrotropic Agents
- Dyes
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
By research - Antibodies
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at info@selleckchem.com to customize your library.
You could select:
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly DYKDDDDK Tag Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- Featured Products
- MRTX1133
- Nab-Paclitaxel
- KP-457
- IAG933
- RMC-6236 (Daraxonrasib)
- RMC-7977
- Zoldonrasib (RMC-9805)
- GsMTx4
- Navitoclax (ABT-263)
- TSA (Trichostatin A)
- Y-27632 Dihydrochloride
- SB431542
- SB202190
- MK-2206 Dihydrochloride
- LY294002
- Alisertib (MLN8237)
- XAV-939
- CHIR-99021 (Laduviglusib)
- Bafilomycin A1 (Baf-A1)
- Thiazovivin (TZV)
- CP-673451
- Verteporfin
- DAPT
- Galunisertib (LY2157299)
- MG132
- SBE-β-CD
- Tween 80
- Bavdegalutamide (ARV-110)
- Z-VAD-FMK
- Wnt-C59 (C59)
- IWR-1-endo
- (+)-JQ1
- 3-Deazaneplanocin A (DZNep) Hydrochloride
- RepSox (E-616452)
- Erastin
- Q-VD-Oph
- Puromycin Dihydrochloride
- Cycloheximide
- Telaglenastat (CB-839)
- A-83-01
- Ceralasertib (AZD6738)
- Liproxstatin-1
- Emricasan (IDN-6556)
- PMA (Phorbol 12-myristate 13-acetate)
- Dibutyryl cAMP (Bucladesine) sodium
- Nedisertib (M3814)
- PLX5622
- IKE (Imidazole Ketone Erastin)
- STM2457
- Saruparib (AZD5305)
- New Products
- Contact Us
research use only
Imatinib PDGFR inhibitor
Cat.No.S2475
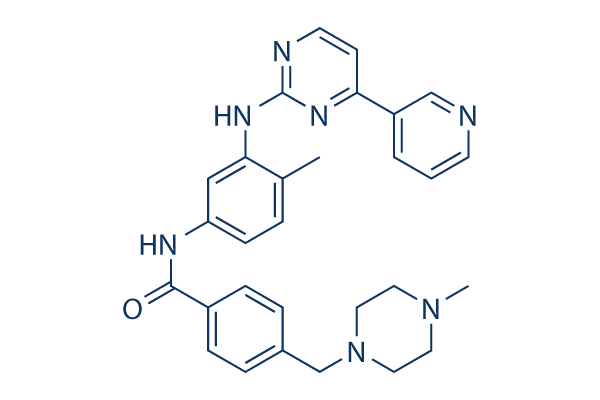
Chemical Structure
Molecular Weight: 493.6
Quality Control
Batch:
Purity:
99.98%
99.98
Cell Culture, Treatment & Working Concentration
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| LAMA-84 | Growth Inhibition Assay | IC50=0.07304 μM | SANGER | |||
| EM-2 | Growth Inhibition Assay | IC50=0.0888 μM | SANGER | |||
| MEG-01 | Growth Inhibition Assay | IC50=0.08921 μM | SANGER | |||
| BV-173 | Growth Inhibition Assay | IC50=0.1874 μM | SANGER | |||
| K-562 | Growth Inhibition Assay | IC50=0.22432 μM | SANGER | |||
| CGTH-W-1 | Growth Inhibition Assay | IC50=0.38374 μM | SANGER | |||
| ST486 | Growth Inhibition Assay | IC50=0.6854 μM | SANGER | |||
| NCI-H1436 | Growth Inhibition Assay | IC50=0.97801 μM | SANGER | |||
| NOS-1 | Growth Inhibition Assay | IC50=1.65383 μM | SANGER | |||
| A498 | Growth Inhibition Assay | IC50=2.57223 μM | SANGER | |||
| BE-13 | Growth Inhibition Assay | IC50=2.62106 μM | SANGER | |||
| SUP-T1 | Growth Inhibition Assay | IC50=3.82907 μM | SANGER | |||
| NCI-H1770 | Growth Inhibition Assay | IC50=5.57262 μM | SANGER | |||
| IMR-5 | Growth Inhibition Assay | IC50=6.22147 μM | SANGER | |||
| LB2241-RCC | Growth Inhibition Assay | IC50=8.07384 μM | SANGER | |||
| TGBC24TKB | Growth Inhibition Assay | IC50=8.34052 μM | SANGER | |||
| SCC-15 | Growth Inhibition Assay | IC50=10.7788 μM | SANGER | |||
| BB49-HNC | Growth Inhibition Assay | IC50=14.3335 μM | SANGER | |||
| ES7 | Growth Inhibition Assay | IC50=14.7379 μM | SANGER | |||
| LB2518-MEL | Growth Inhibition Assay | IC50=16.6094 μM | SANGER | |||
| NCI-H510A | Growth Inhibition Assay | IC50=17.2442 μM | SANGER | |||
| TE-441-T | Growth Inhibition Assay | IC50=17.2886 μM | SANGER | |||
| HH | Growth Inhibition Assay | IC50=17.3999 μM | SANGER | |||
| LC4-1 | Growth Inhibition Assay | IC50=18.0652 μM | SANGER | |||
| KARPAS-45 | Growth Inhibition Assay | IC50=18.1848 μM | SANGER | |||
| LB1047-RCC | Growth Inhibition Assay | IC50=18.4452 μM | SANGER | |||
| NKM-1 | Growth Inhibition Assay | IC50=19.3552 μM | SANGER | |||
| SCLC-21H | Growth Inhibition Assay | IC50=20.1246 μM | SANGER | |||
| RS4-11 | Growth Inhibition Assay | IC50=20.3308 μM | SANGER | |||
| ALL-PO | Growth Inhibition Assay | IC50=20.8149 μM | SANGER | |||
| GDM-1 | Growth Inhibition Assay | IC50=22.5945 μM | SANGER | |||
| DMS-79 | Growth Inhibition Assay | IC50=24.4934 μM | SANGER | |||
| MPP-89 | Growth Inhibition Assay | IC50=25.6874 μM | SANGER | |||
| NB10 | Growth Inhibition Assay | IC50=26.4699 μM | SANGER | |||
| LS-513 | Growth Inhibition Assay | IC50=26.8847 μM | SANGER | |||
| L-540 | Growth Inhibition Assay | IC50=26.9143 μM | SANGER | |||
| ES1 | Growth Inhibition Assay | IC50=27.521 μM | SANGER | |||
| NTERA-S-cl-D1 | Growth Inhibition Assay | IC50=30.5093 μM | SANGER | |||
| EW-1 | Growth Inhibition Assay | IC50=32.9454 μM | SANGER | |||
| Calu-6 | Growth Inhibition Assay | IC50=33.1855 μM | SANGER | |||
| CTV-1 | Growth Inhibition Assay | IC50=33.9789 μM | SANGER | |||
| YT | Growth Inhibition Assay | IC50=38.5209 μM | SANGER | |||
| TE-6 | Growth Inhibition Assay | IC50=41.2798 μM | SANGER | |||
| HT-144 | Growth Inhibition Assay | IC50=41.5486 μM | SANGER | |||
| EW-13 | Growth Inhibition Assay | IC50=42.2791 μM | SANGER | |||
| KALS-1 | Growth Inhibition Assay | IC50=43.1329 μM | SANGER | |||
| MOLT-16 | Growth Inhibition Assay | IC50=45.0752 μM | SANGER | |||
| D-336MG | Growth Inhibition Assay | IC50=45.9599 μM | SANGER | |||
| TE-11 | Growth Inhibition Assay | IC50=46.653 μM | SANGER | |||
| EB2 | Growth Inhibition Assay | IC50=46.699 μM | SANGER | |||
| SK-N-DZ | Growth Inhibition Assay | IC50=48.0961 μM | SANGER | |||
| SW684 | Growth Inhibition Assay | IC50=48.2695 μM | SANGER | |||
| EW-18 | Growth Inhibition Assay | IC50=48.4395 μM | SANGER | |||
| RL95-2 | Growth Inhibition Assay | IC50=50.071 μM | SANGER | |||
| CHP-126 | Growth Inhibition Assay | IC50=50.8905 μM | SANGER | |||
| NCI-H1395 | Growth Inhibition Assay | IC50=51.7835 μM | SANGER | |||
| TE-15 | Growth Inhibition Assay | IC50=52.2556 μM | SANGER | |||
| ES4 | Growth Inhibition Assay | IC50=52.9775 μM | SANGER | |||
| TE-1 | Growth Inhibition Assay | IC50=53.9455 μM | SANGER | |||
| SIMA | Growth Inhibition Assay | IC50=57.3311 μM | SANGER | |||
| LB647-SCLC | Growth Inhibition Assay | IC50=64.1188 μM | SANGER | |||
| KY821 | Growth Inhibition Assay | IC50=64.2552 μM | SANGER | |||
| LC-2-ad | Growth Inhibition Assay | IC50=65.7601 μM | SANGER | |||
| KP-N-RT-BM-1 | Growth Inhibition Assay | IC50=66.6366 μM | SANGER | |||
| SW872 | Growth Inhibition Assay | IC50=67.4382 μM | SANGER | |||
| ES5 | Growth Inhibition Assay | IC50=67.6968 μM | SANGER | |||
| SK-NEP-1 | Growth Inhibition Assay | IC50=68.3803 μM | SANGER | |||
| RPMI-6666 | Growth Inhibition Assay | IC50=71.032 μM | SANGER | |||
| UACC-812 | Growth Inhibition Assay | IC50=71.1609 μM | SANGER | |||
| COLO-829 | Growth Inhibition Assay | IC50=72.6987 μM | SANGER | |||
| KP-N-YS | Growth Inhibition Assay | IC50=72.7139 μM | SANGER | |||
| GI-1 | Growth Inhibition Assay | IC50=73.2868 μM | SANGER | |||
| ETK-1 | Growth Inhibition Assay | IC50=73.4932 μM | SANGER | |||
| LXF-289 | Growth Inhibition Assay | IC50=73.729 μM | SANGER | |||
| CAS-1 | Growth Inhibition Assay | IC50=73.8857 μM | SANGER | |||
| EW-22 | Growth Inhibition Assay | IC50=74.7115 μM | SANGER | |||
| NCI-H2196 | Growth Inhibition Assay | IC50=75.6379 μM | SANGER | |||
| EoL-1-cell | Growth Inhibition Assay | IC50=81.6963 μM | SANGER | |||
| D-247MG | Growth Inhibition Assay | IC50=82.0248 μM | SANGER | |||
| Becker | Growth Inhibition Assay | IC50=82.3481 μM | SANGER | |||
| IST-MEL1 | Growth Inhibition Assay | IC50=82.3482 μM | SANGER | |||
| MDA-MB-134-VI | Growth Inhibition Assay | IC50=82.5996 μM | SANGER | |||
| NCI-H1092 | Growth Inhibition Assay | IC50=84.0197 μM | SANGER | |||
| KINGS-1 | Growth Inhibition Assay | IC50=86.1618 μM | SANGER | |||
| HCC2218 | Growth Inhibition Assay | IC50=86.7913 μM | SANGER | |||
| GI-ME-N | Growth Inhibition Assay | IC50=87.7699 μM | SANGER | |||
| AM-38 | Growth Inhibition Assay | IC50=88.3953 μM | SANGER | |||
| KNS-42 | Growth Inhibition Assay | IC50=89.1018 μM | SANGER | |||
| C8166 | Growth Inhibition Assay | IC50=89.6125 μM | SANGER | |||
| Ramos-2G6-4C10 | Growth Inhibition Assay | IC50=89.8719 μM | SANGER | |||
| CTB-1 | Growth Inhibition Assay | IC50=90.6357 μM | SANGER | |||
| HCE-4 | Growth Inhibition Assay | IC50=91.1336 μM | SANGER | |||
| NCI-H526 | Growth Inhibition Assay | IC50=92.4103 μM | SANGER | |||
| ECC4 | Growth Inhibition Assay | IC50=94.2555 μM | SANGER | |||
| NCCIT | Growth Inhibition Assay | IC50=95.3292 μM | SANGER | |||
| MZ7-mel | Growth Inhibition Assay | IC50=95.904 μM | SANGER | |||
| COLO-684 | Growth Inhibition Assay | IC50=96.2385 μM | SANGER | |||
| SU-DHL-1 | Growth Inhibition Assay | IC50=96.9842 μM | SANGER | |||
| SF126 | Growth Inhibition Assay | IC50=97.5217 μM | SANGER | |||
| NMC-G1 | Growth Inhibition Assay | IC50=98.4554 μM | SANGER | |||
| NB14 | Growth Inhibition Assay | IC50=98.9208 μM | SANGER | |||
| VA-ES-BJ | Growth Inhibition Assay | IC50=99.4056 μM | SANGER | |||
| CHO | Function assay | Inhibition of wild type Platelet-derived growth factor receptor beta phosphorylation in CHO cells, IC50=0.24μM. | 12166950 | |||
| CHO | Function assay | Inhibition of chimeric PDGF receptor with c-kit cytoplasmic domain phosphorylation in CHO cells, IC50=0.26μM. | 12166950 | |||
| CHO | Function assay | Inhibition of chimeric PDGF receptor with CSF-1R cytoplasmic domain phosphorylation in CHO cells, IC50=0.96μM. | 12166950 | |||
| K562 | Cytotoxicity assay | Cytotoxic effect in K562 cells, IC50=0.11μM. | 12951113 | |||
| K562 | Function assay | Inhibitory activity against human K562 cells growth using MTT assay, IC50=0.4μM. | 14552760 | |||
| Sf9 | Function assay | TP_TRANSPORTER: inhibition of ATPase activity in BCRP-expressing Sf9 cells, IC50=0.5μM. | 15155841 | |||
| Sf9 | Function assay | TP_TRANSPORTER: efflux of Hoechst33342 in BCRP-expressing Sf9 cells, IC50=0.9μM. | 15155841 | |||
| K562 | Antiproliferative assay | Antiproliferative activity against K562 cells, IC50=0.182μM. | 16332440 | |||
| U937 | Antiproliferative assay | Antiproliferative activity against U937 cells, IC50=14μM. | 16332440 | |||
| BA/F3 | Antiproliferative assay | 48 hrs | Antiproliferative activity against mouse BA/F3 Bcr-abl negative cells expressing Tel-PDGFRbeta kinase assessed as proliferation after 48 hrs by MTT assay, IC50=0.027μM. | 16415863 | ||
| BA/F3 | Antiproliferative assay | 5 to 10 uM | 48 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Tel-SH2-KD assessed as cell viability at 5 to 10 uM after 48 hrs by MTT assay, IC50=0.055μM. | 16415863 | |
| BA/F3 | Antiproliferative assay | 5 to 10 uM | 48 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Tel-SH3-SH2-KD assessed as cell viability at 5 to 10 uM after 48 hrs by MTT assay, IC50=0.088μM. | 16415863 | |
| 32D | Antiproliferative assay | 48 hrs | Antiproliferative activity against mouse 32D cells transfected with p210 cells expressing Bcr-abl assessed as proliferation after 48 hrs by MTT assay, IC50=0.112μM. | 16415863 | ||
| 32D | Cytotoxicity assay | Cytotoxic activity against mouse 32D cells transfected with p210 Bcr-abl, IC50=0.19μM. | 16415863 | |||
| BA/F3 | Antiproliferative assay | 48 hrs | Antiproliferation activity against mouse BA/F3 cells expressing Bcr-abl assessed as cell viability after 48 hrs by MTT assay, IC50=0.19μM. | 16415863 | ||
| BA/F3 | Antiproliferative assay | 48 hrs | Antiproliferative activity against mouse BA/F3 cells transfected with p210 Bcr-abl assessed as proliferation after 48 hrs by MTT assay, IC50=0.19μM. | 16415863 | ||
| M07e | Antiproliferative assay | 48 hrs | Antiproliferative activity against human M07e Bcr-abl negative cells expressing stem cell factor assessed as proliferation after 48 hrs by MTT assay, IC50=0.257μM. | 16415863 | ||
| BA/F3 | Antiproliferative assay | 48 hrs | Antiproliferation activity against mouse BA/F3 cells expressing Bcr-abl A337N mutant assessed as cell viability after 48 hrs by MTT assay, IC50=0.339μM. | 16415863 | ||
| BA/F3 | Antiproliferative assay | 5 to 10 uM | 48 hrs | Antiproliferative activity against mouse BA/F3 cells expressing NPM-abl assessed as cell viability at 5 to 10 uM after 48 hrs by MTT assay, IC50=0.393μM. | 16415863 | |
| BA/F3 | Antiproliferative assay | 48 hrs | Antiproliferation activity against mouse BA/F3 cells expressing Bcr-abl A334l mutant assessed as cell viability after 48 hrs by MTT assay, IC50=0.962μM. | 16415863 | ||
| 32D | Cytotoxicity assay | Cytotoxic activity against mouse 32D Bcr-abl negative cells, IC50=20μM. | 16415863 | |||
| BaF3 | Antiproliferative assay | Antiproliferative activity against murine BaF3 cells expressing wild type NPM/ALK by [3H]thymidine uptake assay, IC50=7.1μM. | 16970400 | |||
| BaF3 | Antiproliferative assay | Antiproliferative activity against murine BaF3 cells expressing NPM/ALK L256T mutant by [3H]thymidine uptake assay, IC50=9.1μM. | 16970400 | |||
| Vero | Antiviral assay | 3 days | Antiviral activity against Dengue virus infected in african green monkey Vero cells administered before viral challenge after 3 days by viral plaque assay | 17360676 | ||
| Sf9 | Function assay | Inhibition of human Lyn kinase expressed in Sf9 cells, IC50=0.22μM. | 17376680 | |||
| Sf9 | Function assay | Inhibition of human Abl kinase expressed in Sf9 cells, IC50=0.47μM. | 17376680 | |||
| K562 | Cytotoxicity assay | Cytotoxicity against K562 cells by MTT assay, IC50=0.18μM. | 17572088 | |||
| A2780/ADR | Function assay | Inhibition of P-glycoprotein expressed in A2780/ADR cells by calcein AM assay, IC50=7.24436μM. | 17890094 | |||
| MO7e | Function assay | Inhibition of SCF-induced phosphorylation of c-Kit in MO7e cells by TR-FRET assay, IC50=0.046μM. | 18447379 | |||
| MCF7 | Function assay | Inhibition of ABCG2 in human mitoxantrone-resistant MCF7 cells by Hoechst 33342 assay, IC50=4.89779μM. | 18678495 | |||
| A2780 | Function assay | Inhibition of P-gp in human adriamycin-resistant A2780 cells by Hoechst 33342 assay, IC50=7.24436μM. | 18678495 | |||
| CML | Antiproliferative assay | Antiproliferative activity against human CML cells, IC50=0.35μM. | 19219016 | |||
| EOL-1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human EOL-1 cells after 72 hrs by alamar-blue cell viability assay, IC50=0.0001995μM. | 19301902 | ||
| EOL-1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human EOL-1 cells after 72 hrs by alamar-blue cell viability assay, IC50=0.0002μM. | 19301902 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells after 72 hrs by alamar-blue cell viability assay, IC50=0.049μM. | 19301902 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells after 72 hrs by alamar-blue cell viability assay, IC50=0.1μM. | 19301902 | ||
| HCT116 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT116 cells after 72 hrs by alamar-blue cell viability assay, IC50=10μM. | 19301902 | ||
| RKOp21 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human RKOp21 cells after 72 hrs by alamar-blue cell viability assay, IC50=12μM. | 19301902 | ||
| RKOp21 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human RKOp21 cells after 72 hrs by alamar-blue cell viability assay, IC50=12.5892μM. | 19301902 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells after 72 hrs by alamar-blue cell viability assay, IC50=15.5μM. | 19301902 | ||
| MDA-MB-468 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-468 cells after 72 hrs by alamar-blue cell viability assay, IC50=15.8489μM. | 19301902 | ||
| MDA-MB-468 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-468 cells after 72 hrs by alamar-blue cell viability assay, IC50=17μM. | 19301902 | ||
| HeLa | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HeLa cells after 72 hrs by alamar-blue cell viability assay, IC50=18.65μM. | 19301902 | ||
| Hec1A | Cytotoxicity assay | 72 hrs | Cytotoxicity against human Hec1A cells after 72 hrs by alamar-blue cell viability assay, IC50=19μM. | 19301902 | ||
| A549 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A549 cells after 72 hrs by alamar-blue cell viability assay, IC50=19.9526μM. | 19301902 | ||
| CAL27 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CAL27 cells after 72 hrs by alamar-blue cell viability assay, IC50=19.9526μM. | 19301902 | ||
| Hec1A | Cytotoxicity assay | 72 hrs | Cytotoxicity against human Hec1A cells after 72 hrs by alamar-blue cell viability assay, IC50=19.9526μM. | 19301902 | ||
| CAL27 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CAL27 cells after 72 hrs by alamar-blue cell viability assay, IC50=22μM. | 19301902 | ||
| H460 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human H460 cells after 72 hrs by alamar-blue cell viability assay, IC50=24μM. | 19301902 | ||
| PC3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human PC3 cells after 72 hrs by alamar-blue cell viability assay, IC50=24μM. | 19301902 | ||
| HCT15 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT15 cells after 72 hrs by alamar-blue cell viability assay, IC50=25μM. | 19301902 | ||
| HeLa | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HeLa cells after 72 hrs by alamar-blue cell viability assay, IC50=25.1189μM. | 19301902 | ||
| H460 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human H460 cells after 72 hrs by alamar-blue cell viability assay, IC50=25.1189μM. | 19301902 | ||
| HCT15 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT15 cells after 72 hrs by alamar-blue cell viability assay, IC50=25.1189μM. | 19301902 | ||
| PC3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human PC3 cells after 72 hrs by alamar-blue cell viability assay, IC50=25.1189μM. | 19301902 | ||
| Saos2 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human Saos2 cells after 72 hrs by alamar-blue cell viability assay, IC50=25.1189μM. | 19301902 | ||
| Saos2 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human Saos2 cells after 72 hrs by alamar-blue cell viability assay, IC50=26μM. | 19301902 | ||
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MCF7 cells after 72 hrs by alamar-blue cell viability assay, IC50=30μM. | 19301902 | ||
| A431 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A431 cells after 72 hrs by alamar-blue cell viability assay, IC50=31μM. | 19301902 | ||
| H69 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human H69 cells after 72 hrs by alamar-blue cell viability assay, IC50=31.6228μM. | 19301902 | ||
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MCF7 cells after 72 hrs by alamar-blue cell viability assay, IC50=31.6228μM. | 19301902 | ||
| A431 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A431 cells after 72 hrs by alamar-blue cell viability assay, IC50=31.6228μM. | 19301902 | ||
| H69 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human H69 cells after 72 hrs by alamar-blue cell viability assay, IC50=32μM. | 19301902 | ||
| CCRF-CEM | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CCRF-CEM cells after 72 hrs by alamar-blue cell viability assay, IC50=36μM. | 19301902 | ||
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-231 cells after 72 hrs by alamar-blue cell viability assay, IC50=37μM. | 19301902 | ||
| CCRF-CEM/VCR1000 | Cytotoxicity assay | 72 hrs | Cytotoxicity against vincristine-resistant human CCRF-CEM/VCR1000 cells after 72 hrs by alamar-blue cell viability assay, IC50=37μM. | 19301902 | ||
| WM266.4 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human WM266.4 cells after 72 hrs by alamar-blue cell viability assay, IC50=37μM. | 19301902 | ||
| U87MG | Cytotoxicity assay | 72 hrs | Cytotoxicity against human U87MG cells after 72 hrs by alamar-blue cell viability assay, IC50=38μM. | 19301902 | ||
| SKBR3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SKBR3 cells after 72 hrs by alamar-blue cell viability assay, IC50=39μM. | 19301902 | ||
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human MDA-MB-231 cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| SKBR3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human SKBR3 cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| AsPC1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human AsPC1 cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| CCRF-CEM/VCR1000 | Cytotoxicity assay | 72 hrs | Cytotoxicity against vincristine-resistant human CCRF-CEM/VCR1000 cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| CCRF-CEM | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CCRF-CEM cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| U87MG | Cytotoxicity assay | 72 hrs | Cytotoxicity against human U87MG cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| WM266.4 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human WM266.4 cells after 72 hrs by alamar-blue cell viability assay, IC50=39.8107μM. | 19301902 | ||
| AsPC1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human AsPC1 cells after 72 hrs by alamar-blue cell viability assay, IC50=44μM. | 19301902 | ||
| A2780 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human A2780 cells after 72 hrs by alamar-blue cell viability assay, IC50=48μM. | 19301902 | ||
| HT-29 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human HT-29 cells after 96 hrs by SRB assay, IC50=0.06μM. | 19469547 | ||
| GIST882 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human GIST882 cells after 96 hrs by SRB assay, IC50=1.7μM. | 19469547 | ||
| HGC27 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human HGC27 cells after 96 hrs by SRB assay, IC50=2.4μM. | 19469547 | ||
| MCF7 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human MCF7 cells after 96 hrs by SRB assay, IC50=11.5μM. | 19469547 | ||
| GIST48 | Cytotoxicity assay | 3 to 6 days | Cytotoxicity against human GIST48 cells assessed as effect on cell viability after 3 to 6 days by luciferase based luminescence assay, IC50=18.7μM. | 19469547 | ||
| ALL3 | Cytotoxicity assay | Cytotoxicity against Philadelphia chromosome positive human ALL3 cells by Alamar Blue fluorescent assay, IC50=0.333μM. | 19889540 | |||
| MDCK | Function assay | Inhibition of BCRP expressed in MDCK cells by pheophorbide A assay, IC50=3.38μM. | 19932960 | |||
| MCF7 MX | Function assay | Inhibition of BCRP expressed in MCF7 MX cells by Hoechst 33342 staining, IC50=4.07μM. | 19932960 | |||
| KBM5 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human KBM5 cells harboring wild type Bcr-Abl after 72 hrs by MTT assay, IC50=0.28μM. | 20149665 | ||
| KBM5 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human KBM5 cells harboring Bcr-Abl T315I mutant after 72 hrs by MTT assay, IC50=5.4μM. | 20149665 | ||
| FDC-P1 | Function assay | 48 hrs | Inhibition of human FMS expressed in growth factor dependent mouse FDC-P1 cells assessed as inhibition of FMS-mediated cell proliferation in presence human CSF1 after 48 hrs by resazurin dye reduction assay, IC50=1.274μM. | 20156689 | ||
| FDC-P1 | Function assay | 48 hrs | Inhibition of mouse GM-CSF-stimulated cell proliferation of growth factor dependent mouse FDC-P1 cells expressing human FMS after 48 hrs by resazurin dye reduction assay, IC50=8.437μM. | 20156689 | ||
| K562 | Function assay | 2 hrs | Inhibition of BCR/ABL p210 autophosphorylation in human K562 cells after 2 hrs by Western blot analysis, IC50=0.75μM. | 20188579 | ||
| BESM | Antitrypanosomal assay | 88 hrs | Antitrypanosomal activity against Trypanosoma cruzi amastigotes infected in BESM cells measured after 88 hrs postinfection by HTS assay, EC50=9μM. | 20547819 | ||
| BESM | Cytotoxicity assay | 88 hrs | Cytotoxicity against BESM cells after 88 hrs by HTS assay, EC50=32μM. | 20547819 | ||
| BA/F3 | Antiproliferative assay | Antiproliferative activity against PDGFRbeta transfected mouse BA/F3 cells, IC50=0.039μM. | 20817538 | |||
| HEK293 | Function assay | Inhibition of autophosphorylation of DDR1 expressed in HEK293 cells by ELISA, IC50=0.043μM. | 20817538 | |||
| A31 | Function assay | Inhibition of human PDGFRalpha autophosphorylation in human A31 cells by ELISA, IC50=0.072μM. | 20817538 | |||
| A31 | Function assay | Inhibition of PDGFRbeta autophosphorylation in human A31 cells by ELISA, IC50=0.072μM. | 20817538 | |||
| GIST882 | Function assay | Inhibition of human KIT autophosphorylation in human GIST882 cells by ELISA, IC50=0.097μM. | 20817538 | |||
| GIST882 | Antiproliferative assay | Antiproliferative activity against human GIST882 cells, IC50=0.108μM. | 20817538 | |||
| HEK293 | Function assay | Inhibition of autophosphorylation of DDR2 expressed in HEK293 cells by ELISA, IC50=0.141μM. | 20817538 | |||
| Ba/F | Function assay | Inhibition of autophosphorylation of BCR-ABL1 expressed in Ba/F cells, IC50=0.221μM. | 20817538 | |||
| K562 | Antiproliferative assay | Antiproliferative activity against human K562 cells, IC50=0.244μM. | 20817538 | |||
| HEK293 | Function assay | Inhibition of autophosphorylation of CSF1R expressed in HEK293 cells by ELISA, IC50=0.291μM. | 20817538 | |||
| M-NFS-60 | Antiproliferative assay | Antiproliferative activity against mouse M-NFS-60 cells, IC50=0.358μM. | 20817538 | |||
| K562 | Function assay | Inhibition of BCR-ABL1 autophosphorylation in human K562 cells, IC50=0.473μM. | 20817538 | |||
| BA/F3 | Antiproliferative assay | Antiproliferative activity against BCR-ABL1 transfected mouse BA/F3 cells, IC50=0.678μM. | 20817538 | |||
| BA/F3 | Antiproliferative assay | Antiproliferative activity mouse BA/F3 cells in presence of interleukin 3, IC50=9.459μM. | 20817538 | |||
| K562 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human K562 cells after 48 hrs by MTT assay, IC50=0.58μM. | 21295380 | ||
| K562 | Antiproliferative assay | Antiproliferative activity against human imatinib-resistant K562 cells, IC50=0.00605μM. | 21376587 | |||
| KU812 | Antiproliferative assay | Antiproliferative activity against human KU812 cells, IC50=0.337μM. | 21376587 | |||
| K562 | Antiproliferative assay | Antiproliferative activity against human K562 cells, IC50=0.38μM. | 21376587 | |||
| Sf9 | Function assay | Binding affinity to human active site of N-terminal hexahistidine-tagged ABL2 expressed in Trichoplusia ni infected Sf9 cells by isothermal titration calorimetric assay, Kd=0.006μM. | 21417343 | |||
| Sf9 | Function assay | Binding affinity to human remote site of N-terminal hexahistidine-tagged ABL2 expressed in Trichoplusia ni infected Sf9 cells by isothermal titration calorimetric assay, Kd=0.5μM. | 21417343 | |||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=5.4μM. | 21576023 | ||
| Ba/F3 | Function assay | 1 uM | 24 hrs | Inhibition of 14-3-3sigma in human Ba/F3 cells expressing wild type Bcr-Abl construct assessed as release of full-length c-Abl from cytoplasmic complex with 14-3-3sigma at 1 uM after 24 hrs by immunoprecipitation/immunoblot technique | 21962576 | |
| Ba/F3 | Function assay | 1 uM | 24 hrs | Inhibition of 14-3-3sigma in human Ba/F3 cells expressing wild type Bcr-Abl construct assessed as nuclear import of full-length c-Abl at 1 uM after 24 hrs by immunoprecipitation/immunoblot technique | 21962576 | |
| K562 | Function assay | 6 hrs | Inhibition of BCR-ABL-mediated STAT5 phosphorylation in human K562 cells after 6 hrs by phospho-flow cytometry | 22148584 | ||
| K562 | Function assay | 40 uM | 6 hrs | Inhibition of BCR-ABL-mediated ERK1 phosphorylation in human K562 cells at 40 uM after 6 hrs by phospho-flow cytometry | 22148584 | |
| K562 | Function assay | 40 uM | 6 hrs | Inhibition of BCR-ABL-mediated ERK2 phosphorylation in human K562 cells at 40 uM after 6 hrs by phospho-flow cytometry | 22148584 | |
| TF1 | Cytotoxicity assay | Cytotoxicity against human TF1 cells expressing c-KIT mutation assessed as cell viability, IC50=0.013μM. | 22221201 | |||
| primary leukemia cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human primary leukemia cells isolated from acute myeloid leukemia patient expressing wild type FLT3 assessed as cell viability after 72 hrs by luciferase assay, IC50=0.1μM. | 22221201 | ||
| primary leukemia cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human primary leukemia cells isolated from acute myeloid leukemia patient expressing FLT3-D835Y mutation assessed as cell viability after 72 hrs by luciferase assay, IC50=0.5μM. | 22221201 | ||
| K562 | Antiproliferative assay | Antiproliferative activity against human K562 cells, GI50=0.17μM. | 22439674 | |||
| HUVEC | Antiproliferative assay | Antiproliferative activity against HUVEC cells, GI50=18.5μM. | 22439674 | |||
| K562 | Cytotoxicity assay | 24 hrs | Cytotoxicity against human K562 cells after 24 hrs by MTT assay, IC50=0.1μM. | 22632935 | ||
| K562 | Cytotoxicity assay | 48 to 72 hrs | Cytotoxicity against human BCR-ABL positive K562 cells after 48 to 72 hrs by MTT assay, IC50=0.3844μM. | 22789429 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells after 72 hrs by MTS assay, GI50=0.39μM. | 22932313 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl H396P mutant after 72 hrs by CCK-8 assay, IC50=0.11μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl M244V mutant after 72 hrs by CCK-8 assay, IC50=0.129μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl E355G mutant after 72 hrs by CCK-8 assay, IC50=0.362μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl M351T mutant after 72 hrs by CCK-8 assay, IC50=0.424μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl F359V mutant after 72 hrs by CCK-8 assay, IC50=0.96μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl Y253F mutant after 72 hrs by CCK-8 assay, IC50=1.444μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing wild type Bcr-Abl after 72 hrs by CCK-8 assay, IC50=1.834μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl G250E mutant after 72 hrs by CCK-8 assay, IC50=1.863μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl H396R mutant after 72 hrs by CCK-8 assay, IC50=2.377μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl F486S mutant after 72 hrs by CCK-8 assay, IC50=2.547μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl E255K mutant after 72 hrs by CCK-8 assay, IC50=2.951μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl T315I mutant after 72 hrs by CCK-8 assay, IC50=3.005μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl Q252H mutant after 72 hrs by CCK-8 assay, IC50=3.763μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl E255V mutant after 72 hrs by CCK-8 assay, IC50=8.615μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells expressing Bcr-Abl Y253H mutant after 72 hrs by CCK-8 assay, IC50=17.083μM. | 23088644 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells after 72 hrs by CCK-8 assay, IC50=18.52μM. | 23088644 | ||
| MDCK2 | Function assay | 5 mins | Inhibition of human MATE1-mediated [14]-metformin uptake expressed in polarized MDCK2 cells after 5 mins by liquid scintillation counting analysis, IC50=0.04μM. | 23241029 | ||
| HEK293 | Function assay | 1.5 mins | Inhibition of human MATE1-mediated [14]-metformin uptake expressed in HEK293 cells after 1.5 mins by scintillation counting analysis, IC50=0.05μM. | 23241029 | ||
| HEK293 | Function assay | 500 uM | 1.5 mins | Inhibition of human MATE2K-mediated ASP+ uptake expressed in HEK293 cells up to 500 uM after 1.5 mins by fluorescence assay, IC50=0.35μM. | 23241029 | |
| HEK293 | Function assay | 1.5 mins | Inhibition of human MATE1-mediated ASP+ uptake expressed in HEK293 cells after 1.5 mins by fluorescence assay, IC50=0.35μM. | 23241029 | ||
| HEK293 | Function assay | 1.5 mins | Inhibition of human MATE2K-mediated ASP+ uptake expressed in HEK293 cells after 1.5 mins by fluorescence assay, IC50=2.9μM. | 23241029 | ||
| HEK293 | Function assay | 3 mins | Inhibition of human OCT2-mediated ASP+ uptake expressed in HEK293 cells after 3 mins by fluorescence assay, IC50=4.2μM. | 23241029 | ||
| HEK293 | Function assay | 3 mins | Inhibition of human OCT3-mediated ASP+ uptake expressed in HEK293 cells after 3 mins by fluorescence assay, IC50=25.5μM. | 23241029 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant wild type ABL1 expressed in insect cells after 30 mins by FRET assay, IC50=0.0982μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 M351T mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.1143μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 Q252H mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.115μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 H396P mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.1739μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL M351T mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=0.24μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 G250E mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.3599μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 E255K mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.4858μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing wild type BCR-ABL assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=0.5μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL E359V mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=0.59μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 Y253F mutant expressed in insect cells after 30 mins by FRET assay, IC50=0.7496μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL F486S mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=1μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL E355G mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=1.6μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL F317L mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=3.3μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL Q252H mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=3.6μM. | 23301703 | ||
| insect cells | Function assay | 30 mins | Inhibition of human recombinant ABL1 T315I mutant expressed in insect cells after 30 mins by FRET assay, IC50=5.155μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL G250E mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=5.2μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL E255K mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=5.6μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL Y253F mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=9.2μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL F317V mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=9.5μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL H396R mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=11.1μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL T315I mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=12.2μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL L248V mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=12.2μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells expressing BCR-ABL Y253H mutant assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=12.5μM. | 23301703 | ||
| BA/F3 | Cytotoxicity assay | 72 hrs | Cytotoxicity against mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CCK-8 assay, IC50=13.5μM. | 23301703 | ||
| BA/F3 | Antitumor assay | 50 mg/kg | 14 days | Antitumor activity against mouse BA/F3 cells expressing wild type BCR-ABL allografted in SCID mouse assessed as inhibition of tumor growth at 50 mg/kg, po qd for 14 days | 23301703 | |
| K562 | Antiproliferative assay | Antiproliferative activity against human K562 cells, IC50=0.5μM. | 23352483 | |||
| BA/F3 | Cytotoxicity assay | 48 hrs | Cytotoxicity against mouse BA/F3 cells transfected with wild type Bcr-Abl after 48 hrs by XTT assay, IC50=0.092μM. | 23600806 | ||
| K562 | Cytotoxicity assay | Cytotoxicity against human K562 cells, IC50=0.17μM. | 23600806 | |||
| BA/F3 | Cytotoxicity assay | 48 hrs | Cytotoxicity against mouse BA/F3 cells transfected with Bcr-Abl T315I mutant after 48 hrs by XTT assay, IC50=4.79μM. | 23600806 | ||
| Ba/F3 | Function assay | 1 hr | Inhibition of Bcr-Abl T315I mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as reduction of phosphorylated CrkL level after 1 hr by Western blot analysis | 23600806 | ||
| Ba/F3 | Function assay | 1 hr | Inhibition of Bcr-Abl T315I mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as reduction of phosphorylated STAT5 level after 1 hr by Western blot analysis | 23600806 | ||
| Ba/F3 | Function assay | 1 hr | Inhibition of Bcr-Abl T315I mutant (unknown origin) phosphorylation transfected in mouse Ba/F3 cells after 1 hr by Western blot analysis | 23600806 | ||
| GIST882 | Function assay | Inhibition of KIT K642E mutant autophosphorylation (unknown origin) expressed in human GIST882 cells by ELISA, IC50=0.097μM. | 23611771 | |||
| GIST882 | Function assay | Inhibition of KIT K642E mutant (unknown origin) expressed in human GIST882 cells assessed as cell growth inhibition by ATP-depletion assay, IC50=0.108μM. | 23611771 | |||
| 32D | Function assay | Inhibition of human BCR-ABL1 (unknown origin) expressed in mouse 32D cells, IC50=0.194μM. | 23611771 | |||
| BA/F3 | Function assay | Inhibition of human BCR-ABL1 autophosphorylation expressed in mouse BA/F3 cells by ELISA, IC50=0.221μM. | 23611771 | |||
| BA/F3 | Function assay | Inhibition of human BCR-ABL1 expressed in mouse BA/F3 cells assessed as cell growth inhibition by ATP-depletion assay, IC50=0.678μM. | 23611771 | |||
| K562 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human K562 cells assessed as growth inhibition after 48 hrs by MTT assay, IC50=0.47μM. | 23735826 | ||
| GISTT1 | Function assay | 72 hrs | Inhibition of KIT in human GISTT1 cells assessed as growth inhibition after 72 hrs by SRB assay, GI50=0.01μM. | 23773153 | ||
| GIST48B | Growth inhibition assay | 72 hrs | Growth inhibition of human GIST48B cells after 72 hrs by SRB assay, GI50=20μM. | 23773153 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=0.5μM. | 23932071 | ||
| HepG2 | Antiproliferative assay | 24 to 96 hrs | Antiproliferative activity against human HepG2 cells after 24 to 96 hrs by MTS assay, IC50=10μM. | 23932071 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells after 72 hrs by calcein-AM assay, IC50=0.5μM. | 23981532 | ||
| Sf9 | Function assay | Inhibition of recombinant Abl kinase (unknown origin) expressed in Sf9 cells using GGEAIYAAPFKK as substrate in presence of [gamma33P]ATP, IC50=0.3μM. | 24681986 | |||
| K562 | Antiproliferative assay | 24 hrs | In vitro antiproliferative activity against human K562 cells assessed as decrease in cell viability after 24 hrs by MTT assay, IC50=13μM. | 24681986 | ||
| BV173 | Antiproliferative assay | 24 hrs | In vitro antiproliferative activity against human BV173 cells assessed as decrease in cell viability after 24 hrs by MTT assay, IC50=20μM. | 24681986 | ||
| CCRF-CEM | Antiproliferative assay | 24 hrs | In vitro antiproliferative activity against human CCRF-CEM cells assessed as decrease in cell viability after 24 hrs by MTT assay, IC50=45μM. | 24681986 | ||
| K562 | Function assay | 10 uM | 1 hr | Inhibition of Bcr/Abl kinase tyrosine phosphorylation in human K562 cells at 10 uM after 1 hr by immunoblot analysis | 24681986 | |
| K562 | Function assay | 10 uM | 1 hr | Inhibition of Abl kinase tyrosine phosphorylation in human K562 cells at 10 uM after 1 hr by immunoblot analysis | 24681986 | |
| K562 | Function assay | 10 uM | 1 hr | Inhibition of tyrosine phosphorylation of protein with molecular weight between 40 and 60 kDa in human K562 cells at 10 uM after 1 hr by immunoblot analysis | 24681986 | |
| HGC27 | Cytotoxicity assay | 48 hrs | Cytotoxicity against HGC27 cells assessed as cell viability after 48 hrs by MTT assay, IC50=3.8μM. | 24900584 | ||
| HMC-1.2 | Antiproliferative assay | Antiproliferative activity against human HMC-1.2 cells carrying V560G, D816V mutant assessed as cell growth inhibition by MTT assay, IC50=17μM. | 25004409 | |||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against Bcr/Abl positive human K562 cells after 48 hrs by MTT method, IC50=4.12μM. | 25464886 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells after 72 hrs using Calcein AM by fluorescence assay, IC50=0.5μM. | 25757603 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=0.47μM. | 25778766 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells expressing wild type Bcr-Abl after 72 hrs by CCK-8 assay, IC50=0.28μM. | 26195136 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=1.16μM. | 26231079 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against imatinib-resistant human K562 cells after 48 hrs by MTT assay, IC50=15.7μM. | 26231079 | ||
| K562 | Cytotoxicity assay | Cytotoxicity against human K562 cells assessed as reduction in cell viability, IC50=0.06μM. | 26264503 | |||
| IR-K562 | Cytotoxicity assay | Cytotoxicity against imatinib resistant human IR-K562 cells assessed as reduction in cell viability, IC50=1.1μM. | 26264503 | |||
| HEK293 | Cytotoxicity assay | Cytotoxicity against HEK293 cells assessed as reduction in cell viability, CC50=8.4μM. | 26264503 | |||
| U937 | Cytotoxicity assay | Cytotoxicity against human U937 cells assessed as reduction in cell viability, IC50=19μM. | 26264503 | |||
| THP1 | Cytotoxicity assay | Cytotoxicity against human THP1 cells assessed as reduction in cell viability, IC50=25μM. | 26264503 | |||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=4.12μM. | 26298495 | ||
| HCT116 | Anticancer assay | Anticancer activity against human HCT116 cells, IC50=34.4μM. | 26312434 | |||
| K562 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human K562 cells expressing Bcr-Abl assessed as growth inhibition after 48 hrs by MTT assay, IC50=1.16μM. | 26451772 | ||
| BA/F3 | Growth inhibition assay | 48 hrs | Growth inhibition of mouse BA/F3 cells expressing wild-type Bcr-Abl after 48 hrs by MTT assay, IC50=0.089μM. | 26562217 | ||
| BA/F3 | Growth inhibition assay | 48 hrs | Growth inhibition of mouse BA/F3 cells expressing Bcr-Abl T315I mutant after 48 hrs by MTT assay, IC50=14.5μM. | 26562217 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by trypan blue dye exclusion assay, IC50=0.22μM. | 26629859 | ||
| K562 | Apoptosis assay | 48 hrs | Induction of apoptosis in human K562 cells after 48 hrs by annexinV and propidium iodide staining based flow cytometry, AC50=0.68μM. | 26629859 | ||
| K562 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human K562 cells after 48 hrs by MTT assay, IC50=0.53μM. | 26707846 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells assessed as cell viability after 72 hrs by fluorescence microplate reader method, IC50=0.73μM. | 26741853 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells assessed as cell viability after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.14μM. | 26789553 | ||
| KU812 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human KU812 cells assessed as cell viability after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.16μM. | 26789553 | ||
| MEG01 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MEG01 cells assessed as cell viability after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.24μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210 (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.38μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-LCK (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.5μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-M351T mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.625μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-Q252H mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.659μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-F317I mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.855μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-H369P mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=1.69μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-E255K mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=1.93μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-SRC (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=2.1μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210-F317L mutant (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=2.169μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-DDR1 (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=3μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-BLK (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=4.1μM. | 26789553 | ||
| HEL | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HEL cells assessed as cell viability after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=5.3μM. | 26789553 | ||
| BA/F3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BA/F3 cells assessed as cell viability after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=6.7μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-DDR2 (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=7.7μM. | 26789553 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-HCK (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=9.7μM. | 26789553 | ||
| Neuro2a | Function assay | 1 uM | Inhibition of DR24 in Dhcr7-deficient mouse Neuro2a cells assessed as decrease in 7-DHC levels at 1 uM by LC-MS/GC-MS analysis | 26789657 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells assessed as cell viability after 72 hrs by MTT assay, IC50=0.51μM. | 26814890 | ||
| K562/G | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562/G cells assessed as cell viability after 72 hrs by MTT assay, IC50=4.65μM. | 26814890 | ||
| BA/F3 | Function assay | 48 hrs | Inhibition of imatinib-resistant PDGFR D842V mutant (unknown origin) expressed in mouse BA/F3 cells assessed as decrease in cell viability after 48 hrs by CellTiter 96 aqueous one solution cell proliferation assay, IC50=0.642μM. | 26844689 | ||
| HL60 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HL60 cells assessed as inhibition of cell survival after 72 hrs by MTT assay, IC50=0.03μM. | 26850004 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells assessed as inhibition of cell survival after 72 hrs by MTT assay, IC50=0.38μM. | 26850004 | ||
| KG1a | Antiproliferative assay | 72 hrs | Antiproliferative activity against human KG1a cells assessed as inhibition of cell survival after 72 hrs by MTT assay, IC50=16.7μM. | 26850004 | ||
| 293T | Cytotoxicity assay | Cytotoxicity against human 293T cells assessed as growth inhibition, CC50=20.53μM. | 26850004 | |||
| BAF3 | Function assay | 72 hrs | Inhibition of human wild type BCR-ABL expressed in mouse BAF3 cells assessed as inhibition of cell proliferation after 72 hrs by MTT assay, EC50=0.034μM. | 27010810 | ||
| JURL-MK1 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human JURL-MK1 cells after 48 hrs by Cell-Titer-Glo luminescent cell viability assay, GI50=0.06μM. | 27011159 | ||
| BV173 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human BV173 cells after 48 hrs by Cell-Titer-Glo luminescent cell viability assay, GI50=0.1μM. | 27011159 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by Cell-Titer-Glo luminescent cell viability assay, GI50=0.2μM. | 27011159 | ||
| EM2 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human EM2 cells after 48 hrs by Cell-Titer-Glo luminescent cell viability assay, GI50=0.26μM. | 27011159 | ||
| KCL22 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human KCL22 cells after 48 hrs by Cell-Titer-Glo luminescent cell viability assay, GI50=0.43μM. | 27011159 | ||
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against cKIT dependent human GISTT1 cells assessed as reduction in cell viability after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.008μM. | 27077705 | ||
| GIST882 | Antiproliferative assay | 72 hrs | Antiproliferative activity against cKIT dependent human GIST882 cells assessed as reduction in cell viability after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.014μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT V559D mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.039μM. | 27077705 | ||
| MEG01 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human BCR-ABL dependent MEG01 cells assessed as reduction in cell viability after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.074μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT L567P mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.1μM. | 27077705 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against BCR-ABL dependent human K562 cells assessed as reduction in cell viability after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.12μM. | 27077705 | ||
| KU812 | Antiproliferative assay | 72 hrs | Antiproliferative activity against BCR-ABL dependent human KU812 cells assessed as reduction in cell viability after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.16μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=0.4μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT N822K mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=1.29μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT V654A mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=2.49μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT V559D/V654A double mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=3μM. | 27077705 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused cKIT T670I/V559D double mutant (unknown origin) transfected in mouse BAF3 cells assessed as decrease in cell proliferation after 72 hrs by celltiter-glo/CCK8 assay, GI50=6.67μM. | 27077705 | ||
| K562 | Cytotoxicity assay | Cytotoxicity against human K562 cells, IC50=0.5μM. | 27189674 | |||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells assessed as decrease in cell proliferation after 72 hrs by XTT assay, IC50=0.31μM. | 27214512 | ||
| A10 | Function assay | 68 hrs | Inhibition of PDGFR-beta driven proliferation of rat A10 cells after 68 hrs in presence of rat recombinant PDGF-BB by cell titer-glo luminescence assay, IC50=0.162μM. | 27502700 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT L567P mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.01μM. | 27545040 | ||
| GIST-T1 | Function assay | 72 hrs | Inhibition of wild type cKIT in human GIST-T1 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.027μM. | 27545040 | ||
| Sf9 | Function assay | 1 hr | Inhibition of His-tagged cKIT (unknown origin) expressed in Sf9 cells using Poly(4:1 Glu, Tyr) peptide as substrate after 1 hr by ADP-glo kinase assay, IC50=0.319μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT N822K mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.36μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.59μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT V654A mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.7μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT V559D/V654A double mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.7μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT T670I/V559D double mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=5.2μM. | 27545040 | ||
| GIST-5R | Function assay | 72 hrs | Inhibition of wild type cKIT T670I mutant in human GIST-5R cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=8.3μM. | 27545040 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT T670I mutant (unknown origin) expressed in mouse BA/F3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=9.6μM. | 27545040 | ||
| Sf9 | Function assay | 1 hr | Inhibition of His-tagged cKIT T670I mutant (544 to 935 residues) (unknown origin) expressed in Sf9 cells using Poly(4:1 Glu, Tyr) peptide as substrate after 1 hr by ADP-glo kinase assay, IC50=9.818μM. | 27545040 | ||
| K562 | Function assay | 100 uM | 8 hrs | Inhibition of BCR-ABL signaling pathway in human K562 cells assessed as suppression of STAT5 phosphorylation at 100 uM after 8 hrs in presence of MeBS by Western blot analysis | 27666635 | |
| K562 | Function assay | 100 uM | 8 hrs | Inhibition of BCR-ABL in signaling pathway human K562 cells assessed as suppression of CrkL phosphorylation at 100 uM after 8 hrs in presence of MeBS by Western blot analysis | 27666635 | |
| K562 | Function assay | 100 uM | 8 hrs | Inhibition of BCR-ABL phosphorylation in human K562 cells at 100 uM after 8 hrs in presence of MeBS by Western blot analysis | 27666635 | |
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GISTT1 cells measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.008μM. | 27966954 | ||
| GIST882 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GIST882 cells measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.014μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused PDGFR-beta (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.019μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused PDGFR-alpha (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.034μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT V559D mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.039μM. | 27966954 | ||
| MEG01 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MEG01 cells measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.074μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT L576P mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.102μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused CSF1R (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.11μM. | 27966954 | ||
| KU812 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human KU812 cells measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.163μM. | 27966954 | ||
| SF9 | Function assay | 1 hr | Inhibition of C-terminal His-tagged human ABL1 expressed in baculovirus infected SF9 cells using Tyr 02 peptide as substrate measured after 1 hr by FRET based Z'Lyte assay, IC50=0.223μM. | 27966954 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.267μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of human BCR/ABL p210 fusion protein expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.27μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.37μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210 fusion protein M356T mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.625μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210 fusion protein F317I mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=0.85μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT N822K mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=1.29μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210 fusion protein H369P mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=1.79μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL p210 fusion protein F317L mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=2.16μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT V654A mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=2.49μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT V559D/V654A double mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=3μM. | 27966954 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of Tel-fused c-KIT T670I mutant (unknown origin) expressed in BAF3 cells assessed as growth inhibition measured after 72 hrs by CellTiter-Glo or CCK8 assay, GI50=6.67μM. | 27966954 | ||
| K562 | Cell cycle assay | 1 uM | 24 hrs | Cell cycle arrest in human K562 cells assessed as accumulation at G0/G1 phase at 1 uM after 24 hrs by propidium iodide/RNase staining based flow cytometry | 27966954 | |
| KU812 | Cell cycle assay | 1 uM | 12 hrs | Cell cycle arrest in human KU812 cells assessed as accumulation at G0/G1 phase at 1 uM after 12 hrs by propidium iodide/RNase staining based flow cytometry | 27966954 | |
| MEG01 | Cell cycle assay | 1 uM | 24 hrs | Cell cycle arrest in human MEG01 cells assessed as accumulation at G0/G1 phase at 1 uM after 24 hrs by propidium iodide/RNase staining based flow cytometry | 27966954 | |
| GISTT1 | Cell cycle assay | 1 uM | 24 hrs | Cell cycle arrest in human GISTT1 cells assessed as accumulation at G0/G1 phase at 1 uM after 24 hrs by propidium iodide/RNase staining based flow cytometry | 27966954 | |
| GIST882 | Cell cycle assay | 1 uM | 48 hrs | Cell cycle arrest in human GIST882 cells assessed as accumulation at G0/G1 phase at 1 uM after 48 hrs by propidium iodide/RNase staining based flow cytometry | 27966954 | |
| K562 | Apoptosis assay | 1 uM | Induction of apoptosis in human K562 cells assessed as cleavage of PARP at 1 uM by Western blot analysis | 27966954 | ||
| K562 | Apoptosis assay | 1 uM | Induction of apoptosis in human K562 cells assessed as increase in cleaved caspase-3 level at 1 uM by Western blot analysis | 27966954 | ||
| KU812 | Apoptosis assay | 1 uM | Induction of apoptosis in human KU812 cells assessed as cleavage of PARP at 1 uM by Western blot analysis | 27966954 | ||
| KU812 | Apoptosis assay | 1 uM | Induction of apoptosis in human KU812 cells assessed as increase in cleaved caspase-3 level at 1 uM by Western blot analysis | 27966954 | ||
| MEG01 | Apoptosis assay | 1 uM | Induction of apoptosis in human MEG01 cells assessed as increase in cleaved caspase-3 level at 1 uM by Western blot analysis | 27966954 | ||
| GISTT1 | Apoptosis assay | 1 uM | Induction of apoptosis in human GISTT1 cells assessed as increase in cleaved caspase-3 level at 1 uM by Western blot analysis | 27966954 | ||
| MEG01 | Apoptosis assay | 1 uM | Induction of apoptosis in human MEG01 cells assessed as cleavage of PARP at 1 uM by Western blot analysis | 27966954 | ||
| GISTT1 | Apoptosis assay | 1 uM | Induction of apoptosis in human GISTT1 cells assessed as cleavage of PARP at 1 uM by Western blot analysis | 27966954 | ||
| GIST882 | Apoptosis assay | 1 uM | Induction of apoptosis in human GIST882 cells assessed as cleavage of PARP at 1 uM by Western blot analysis | 27966954 | ||
| GIST882 | Apoptosis assay | 1 uM | Induction of apoptosis in human GIST882 cells assessed as increase in cleaved caspase-3 level at 1 uM by Western blot analysis | 27966954 | ||
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in AKT phosphorylation at S473 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y703 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y719 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y823 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y703 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y719 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT autophosphorylation at Y823 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in AKT phosphorylation at S473 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in ERK1/2 phosphorylation at T202/Y204 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in Stat3/5 phosphorylation at Y705/Y694 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in S6 phosphorylation at S235/236 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GISTT1 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in S6K phosphorylation at T389 in human GISTT1 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in Stat3/5 phosphorylation at Y705/Y694 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in ERK1/2 phosphorylation at T202/Y204 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in S6 phosphorylation at S235/236 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| GIST882 | Function assay | 1 uM | 2 hrs | Inhibition of c-KIT mediated signaling pathways assessed as reduction in S6K phosphorylation at T389 in human GIST882 cells at 1 uM measured after 2 hrs by immunoblotting | 27966954 | |
| K562 | Function assay | 30 mins | Inhibition of kinobead binding to NQO2 in human K562 cells incubated for 30 mins by iTRAQ reagent-based mass spectrometric method, IC50=0.043μM. | 28280261 | ||
| K562 | Function assay | 30 mins | Inhibition of kinobead binding to DDR1 in human K562 cells incubated for 30 mins by iTRAQ reagent-based mass spectrometric method, IC50=0.09μM. | 28280261 | ||
| K562 | Function assay | 30 mins | Inhibition of kinobead binding to ABL in human K562 cells incubated for 30 mins by iTRAQ reagent-based mass spectrometric method, IC50=0.25μM. | 28280261 | ||
| K562 | Function assay | 30 mins | Inhibition of kinobead binding to ARG in human K562 cells incubated for 30 mins by iTRAQ reagent-based mass spectrometric method, IC50=0.272μM. | 28280261 | ||
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human K562 cells after 48 hrs by MTT assay, IC50=0.53μM. | 28525838 | ||
| A549 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human A549 cells after 48 hrs by MTT assay, IC50=4.56μM. | 28525838 | ||
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GISTT1 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.003μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused PDGFRalpha (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.0107μM. | 28541695 | ||
| GIST882 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GIST882 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.02μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT C674S mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.0249μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT V559D mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.036μM. | 28541695 | ||
| MEG01 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MEG01 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.045μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT L576P mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.0451μM. | 28541695 | ||
| KU812 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human KU812 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.11μM. | 28541695 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.147μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT A829P mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.1482μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.3819μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT V559D/V654A double mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.803μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT V654A mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=0.8031μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT D816H mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=1.245μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL-fused cKIT N822K mutant (unknown origin) expressed in mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=1.466μM. | 28541695 | ||
| BAF3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against mouse BAF3 cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=4.686μM. | 28541695 | ||
| GIST48B | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GIST48B cells assessed as cell growth inhibition after 72 hrs by cell titer-glo assay, GI50=6.986μM. | 28541695 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of ABL in mouse BAF3 cells assessed as reduction in cell viability after 72 hrs by MTT assay, IC50=0.73μM. | 28974338 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of ABL T315I mutant in mouse BAF3 cells assessed as reduction in cell viability after 72 hrs by MTT assay, IC50=20.03μM. | 28974338 | ||
| GISTT1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human GISTT1 cells assessed as cell growth inhibition after 72 hrs by CellTiterGlo assay, GI50=0.04μM. | 28991465 | ||
| GIST430 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human GIST430 cells harboring KIT V654A mutant assessed as cell growth inhibition after 72 hrs by CellTiterGlo assay, GI50=1.1μM. | 28991465 | ||
| Sf9 | Function assay | Binding affinity to recombinant full length human N-terminal GST-tagged Syk-KD (356 to 635 residues) cytoplasmic domain expressed in baculovirus infected Sf9 insect cells using poly EY 4:1 as substrate, Ki=5μM. | 29132752 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for DAOY cells | 29435139 | |||
| EOL-1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human EOL-1 cells after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50<0.001μM. | 29544149 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused PDGFRbeta (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.019μM. | 29544149 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused PDGFRalpha (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.034μM. | 29544149 | ||
| NCI-H1703 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human NCI-H1703 cells after 72 hrs in presence of PDGF-AA by CellTiter-Glo or CCK-8 assay, GI50=0.23μM. | 29544149 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused c-KIT (unknown origin) expressed in mouse BA/F3 cells assessed as growth inhibition after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=0.37μM. | 29544149 | ||
| NCI-H1703 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human NCI-H1703 cells after 72 hrs by CellTiter-Glo or CCK-8 assay, GI50=2.1μM. | 29544149 | ||
| NCI-H1703 | Function assay | 1 uM | 2 hrs | Inhibition of PDGF-induced PDGFRalpha autophosphorylation at Y1018 residue in human NCI-H1703 cells at 1 uM after 2 hrs in presence of PDGF-AA by Western blot analysis | 29544149 | |
| NCI-H1703 | Function assay | 1 uM | 2 hrs | Inhibition of PDGF-induced PDGFRalpha autophosphorylation at Y754 residue in human NCI-H1703 cells at 1 uM after 2 hrs in presence of PDGF-AA by Western blot analysis | 29544149 | |
| NCI-H1703 | Function assay | 1 uM | 2 hrs | Inhibition of PDGF-induced PDGFRalpha autophosphorylation at Y849 residue in human NCI-H1703 cells at 1 uM after 2 hrs in presence of PDGF-AA by Western blot analysis | 29544149 | |
| EOL-1 | Function assay | 1 uM | 2 hrs | Inhibition of PDGFRalpha in human EOL-1 cells assessed as decrease in STAT5 phosphorylation at Y694 residue at 1 uM after 2 hrs in presence of PDGF-AA by Western blot analysis | 29544149 | |
| EOL-1 | Function assay | 1 uM | 2 hrs | Inhibition of PDGFRalpha in human EOL-1 cells assessed as decrease in ERK phosphorylation at T202/Y204 residues at 1 uM after 2 hrs in presence of PDGF-AA by Western blot analysis | 29544149 | |
| K562 | Cytotoxicity assay | 48 hrs | Cytotoxicity in drug sensitive human K562 cells assessed as reduction cell viability incubated for 48 hrs by XTT assay, IC50=1.1μM. | 29655981 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells after 72 hrs by MTT assay, IC50=3.43μM. | 29684708 | ||
| MCF7 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human MCF7 cells after 72 hrs by MTT assay, IC50=7.12μM. | 29684708 | ||
| A2780 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A2780 cells after 72 hrs by MTT assay, IC50=8.9μM. | 29684708 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by MTT assay, IC50=19.66μM. | 29684708 | ||
| HT-29 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human HT-29 cells after 96 hrs by SRB assay, IC50=0.06μM. | 29724653 | ||
| GIST882 | Antiproliferative assay | 96 hrs | Antiproliferative activity against imatinib-susceptible human GIST882 cells harboring c-KIT K642E mutant after 96 hrs by SRB assay, IC50=1.7μM. | 29724653 | ||
| HGC27 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human HGC27 cells after 96 hrs by SRB assay, IC50=2.4μM. | 29724653 | ||
| MCF7 | Antiproliferative assay | 96 hrs | Antiproliferative activity against human MCF7 cells after 96 hrs by SRB assay, IC50=11.3μM. | 29724653 | ||
| GIST48 | Antiproliferative assay | 96 hrs | Antiproliferative activity against imatinib-resistant human GIST48 cells harboring c-KIT V560D/D820A double mutant after 96 hrs by SRB assay, IC50=19.8μM. | 29724653 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 G250H mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.0771μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of wild type BCR-ABL1 (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.0905μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 E355G mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.149μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 E459K mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.201μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 F359V mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.249μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 Q252H mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.455μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 Y253H mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.836μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 E255K mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.838μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 E255V mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of BCR-ABL1-mediated cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=0.874μM. | 30137981 | ||
| Ba/F3 | Function assay | 48 hrs | Inhibition of BCR-ABL1 T315I mutant (unknown origin) expressed in mouse Ba/F3 cells assessed as inhibition of cell proliferation after 48 hrs by BriteLight luciferase assay, GI50=9.645μM. | 30137981 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.017μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused PDGFRalpha (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.046μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit V560D mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.07μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of TEL-fused PDGFRbeta (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.102μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 9 AY502 to 503 insertion mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.192μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and D820A mutant and D820A mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.216μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and N822K mutant and Y823D mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.223μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and A829P mutant and Y823D mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.312μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and V654A mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.392μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and D816H mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.535μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of PDGFRalpha V561D/D842V mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.567μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (557 to 558 residues) and Y823D mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.669μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit V560D/V654A mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=0.701μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit V560D/D816H mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=1.191μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 11 deletion (560 to 578 residues) mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=1.523μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 9 AY502 to 503 insertion and D816 mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=2.749μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit exon 9 AY502 to 503 insertion and V654 mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=3.947μM. | 30204441 | ||
| BA/F3 | Function assay | 72 hrs | Inhibition of Kit D816V mutant (unknown origin) transfected in mouse BA/F3 cells assessed as inhibition of cell growth incubated for 72 hrs by MTS assay, GI50=9.93μM. | 30204441 | ||
| K562 | Cytotoxicity assay | 24 hrs | Cytotoxicity against imatinib-sensitive human K562 cells assessed as decrease in cell viability after 24 hrs by XTT assay, IC50=1μM. | 30261468 | ||
| K562 | Function assay | 1 uM | Inhibition of BCR-ABL in imatinib-sensitive human K562 cells assessed as decrease in CrKL phosphorylation at 1 uM by immuno blot analysis | 30261468 | ||
| Sf9 | Function assay | 60 mins | Inhibition of inactive wild type His-tagged ABL (229 to 500 residues) (unknown origin) expressed in baculovirus infected sf9 cells assessed as reduction in autophosphorylation preincubated for 60 mins followed by ATP addition measured after 8 hrs by ADP-G, IC50=0.043μM. | 30317026 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity in human K562 cells after 72 hrs by CCK8 assay, GI50=0.25μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of wild type C-terminal FLAG-tagged human TEL fused ABL (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.26μM. | 30317026 | ||
| MEG01 | Antiproliferative assay | 72 hrs | Antiproliferative activity in human MEG01 cells after 72 hrs by CCK8 assay, GI50=0.28μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of wild type BCR/ABL (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.29μM. | 30317026 | ||
| KU812 | Antiproliferative assay | 72 hrs | Antiproliferative activity in human KU812 cells after 72 hrs by CCK8 assay, GI50=0.31μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL V299L mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.65μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL M351T mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.85μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL F317I mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.95μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL Q252H mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=1.17μM. | 30317026 | ||
| Sf9 | Function assay | 1 hr | Inhibition of active wild type His-tagged ABL (229 to 500 residues) (unknown origin) expressed in baculovirus infected sf9 cells using ABLtide as substrate after 1 hr by ADP-Glo assay, IC50=1.5μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL F317L mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=2.24μM. | 30317026 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of BCR/ABL H369P mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=3.75μM. | 30317026 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against p53+/+ human HCT116 cells after 72 hrs by CellTiter 96 aqueous one solution assay, IC50=44.55μM. | 30562697 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells after 72 hrs by MTT assay, IC50=0.00015μM. | 30605831 | ||
| MCF7 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MCF7 cells assessed as reduction in cell viability incubated for 48 hrs by MTT assay, IC50=6.33μM. | 30798049 | ||
| MDA-MB-453 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human MDA-MB-453 cells assessed as reduction in cell viability incubated for 48 hrs by MTT assay, IC50=12.84μM. | 30798049 | ||
| GIST882 | Antiproliferative assay | Antiproliferative activity against human GIST882 cells harboring c-KIT exon 13 homozygous primary K642E mutant assessed as reduction in cell viability by MTS assay, GI50=0.195μM. | 30968693 | |||
| GIST882 | Antiproliferative assay | Antiproliferative activity against human GIST882 cells harboring c-KIT exon 13 homozygous primary K642E mutant assessed as reduction in cell viability by methylene blue staining based assay, GI50=0.195μM. | 30968693 | |||
| GIST48 | Antiproliferative assay | Antiproliferative activity against human GIST48 cells harboring c-KIT exon 11 homozygous primary missense V560D mutant and exon 17 heterozygous secondary D820A mutant assessed as reduction in cell viability by MTS assay, GI50=0.625μM. | 30968693 | |||
| GIST48 | Antiproliferative assay | Antiproliferative activity against human GIST48 cells harboring c-KIT exon 11 homozygous primary missense V560D mutant and exon 17 heterozygous secondary D820A mutant assessed as reduction in cell viability by methylene blue staining based assay, GI50=0.625μM. | 30968693 | |||
| Sf9 | Function assay | Inhibition of unactivated recombinant human N-terminal 6x-His-tagged c-KIT (547 to 935 residues)/(694 to 753 residues deletion) expressed in baculovirus infected Sf9 insect cells assessed as decrease in poly (Glu,Tyr) 4:1 phosphorylation, IC50=0.769μM. | 30968693 | |||
| GIST430 | Antiproliferative assay | Antiproliferative activity against human GIST430 cells harboring c-KIT exon 11 primary in-frame V560-L576 deletion mutant and exon 13 heterozygous secondary missense V654A mutant assessed as reduction in cell viability by MTS assay, GI50=0.929μM. | 30968693 | |||
| GIST430 | Antiproliferative assay | Antiproliferative activity against human GIST430 cells harboring c-KIT exon 11 primary in-frame V560-L576 deletion mutant and exon 13 heterozygous secondary missense V654A mutant assessed as reduction in cell viability by methylene blue staining based assay, GI50=0.929μM. | 30968693 | |||
| Sf9 | Function assay | Inhibition of ATP-activated recombinant human N-terminal 6x-His-tagged c-KIT (547 to 935 residues)/(694 to 753 residues deletion) expressed in baculovirus infected Sf9 insect cells assessed as decrease in poly (Glu,Tyr) 4:1 phosphorylation, IC50=5.21μM. | 30968693 | |||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT V559G mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.001μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT V559A mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.005μM. | 31046271 | ||
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GISTT1 cells harboring c-KIT 560 to 578 deletion mutant incubated for 72 hrs by CCK8 assay, GI50=0.026μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT V559D mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.039μM. | 31046271 | ||
| GIST882 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GIST882 cells harboring c-KIT K642E mutant incubated for 72 hrs by CCK8 assay, GI50=0.058μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT L576P mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.115μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT D820E mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.267μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of wild type TEL fused c-KIT (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.364μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT A829P mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.406μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT D816H mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.823μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT N822K mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=1.29μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT V654A/V559D double mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=1.57μM. | 31046271 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of TEL fused c-KIT V654A mutant (unknown origin) transfected in mouse BAF3 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=2.3μM. | 31046271 | ||
| K562 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human K562 cells assessed as inhibition of cell viability incubated for 72 hrs by real time live-cell imaging technique, IC50=0.14μM. | 31097376 | ||
| K562 | Apoptosis assay | 10 uM | 48 hrs | Induction of apoptosis in human K562 cells assessed as increase in apoptotic cells at 10 uM incubated for 48 hrs by Caspase-3/7 reagent-based fluorescence assay | 31097376 | |
| K562 | Antiproliferative assay | 48 hrs | Antiproliferative activity against human BCR/ABL positive K562 cells assessed as growth inhibition measured after 48 hrs by MTT assay, IC50=4.26μM. | 31185413 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V560D mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.003μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V559G mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.008μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V559D mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.01μM. | 31250638 | ||
| GISTT1 | Function assay | 72 hrs | Inhibition of c-KIT 560 to 578 deletion mutant in human GISTT1 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.015μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT D820E mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.035μM. | 31250638 | ||
| GIST882 | Function assay | 72 hrs | Inhibition of c-KIT K642E mutant in human GIST882 cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.048μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT L576P mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.085μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT S709F mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.115μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT D816E mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.174μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT D820G mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.337μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.35μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT D820Y mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.435μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT A829P mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.58μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V654A/V559D double mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.608μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT D816H mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.651μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V559A mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=0.905μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT V654A mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=1.41μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT N822K mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=1.47μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT T670E mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=4.08μM. | 31250638 | ||
| BAF3 | Function assay | 72 hrs | Inhibition of c-KIT Y823D mutant in mouse BAF3(transformed) cells assessed as growth inhibition after 72 hrs by CCK8 assay, GI50=5.87μM. | 31250638 | ||
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GISTT1 cells harboring c-KIT 560 to 578 deletion mutant measured after 72 hrs by CCK8 assay | 31250638 | ||
| EOL-1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against FIP1L1-PDGFRA positive human EOL-1 cells assessed as growth inhibition after 72 hrs by resazurin dye-based fluorescence analysis, GI50<0.0005μM. | 31514019 | ||
| K562 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human K562 cells assessed as growth inhibition after 72 hrs by resazurin dye-based fluorescence analysis, GI50=0.73μM. | 31514019 | ||
| CEM | Cytotoxicity assay | 72 hrs | Cytotoxicity against human CEM cells assessed as growth inhibition after 72 hrs by resazurin dye-based fluorescence analysis, GI50=35.809μM. | 31514019 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of wild type BCR/Abl (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.1438μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl P230 mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.2215μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl P190 mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.3455μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl F317I mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.364μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl V299L mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.4053μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl Q252H mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.4865μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl F317L mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.7163μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl M351T mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=0.7993μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl H396P mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=2.706μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl E255K mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=3.257μM. | 31699612 | ||
| Ba/F3 | Function assay | 72 hrs | Inhibition of BCR/Abl Y253F mutant (unknown origin) transfected in mouse Ba/F3 cells assessed as inhibition of cell proliferation measured after 72 hrs by CellTiter-Glo luminescent cell viability assay, IC50=8.778μM. | 31699612 | ||
| GISTT1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human GISTT1 cells harboring heterozygous deletion mutation at C-kit exon 11 assessed as cell growth inhibition after 72 hrs by CellTiter 96 AQueous One Solution Cell Proliferation assay, GI50=0.04μM. | 31721578 | ||
| Sf9 | Function assay | 150 mins | Inhibition of recombinant N-terminal 6x-His-tagged c-KIT (547 to 935 residues)/(694 to 753 residues deletion) (unknown origin) expressed in baculovirus infected Sf9 insect cells using poly (Glu,Tyr) 4:1 as substrate measured after 150 mins in presence of , IC50=0.053μM. | 31721578 | ||
| Sf9 | Function assay | 4 hrs | Inhibition of wild type recombinant GST-tagged FLT3 (Y567 to S993 residues) (unknown origin) expressed in baculovirus infected Sf9 insect cells using Her2 peptide as substrate measured after 4 hrs in presence of ATP by Kinase-Glo Plus reagent-based lumine, IC50=0.131μM. | 31721578 | ||
| GIST882 | Antiproliferative assay | 144 hrs | Antiproliferative activity against human GIST882 cells harboring c-KIT exon 13 homozygous primary K642E mutant assessed as cell growth inhibition after 144 hrs by methylene blue staining based assay, GI50=0.195μM. | 31721578 | ||
| GIST430 | Antiproliferative assay | 120 hrs | Antiproliferative activity against human GIST430 cells harboring c-KIT exon 11 primary in-frame V560-L576 deletion mutant and exon 13 heterozygous secondary missense V654A mutant assessed as cell growth inhibition after 120 hrs by methylene blue staining , GI50=0.625μM. | 31721578 | ||
| GIST48 | Antiproliferative assay | 120 hrs | Antiproliferative activity against human GIST48 cells harboring c-KIT exon 11 homozygous primary missense V560D mutant and exon 17 heterozygous secondary D820A mutant assessed as cell growth inhibition after 120 hrs by methylene blue staining based assay, GI50=0.929μM. | 31721578 | ||
| Click to View More Cell Line Experimental Data | ||||||
Chemical Information, Storage & Stability
| Molecular Weight | 493.6 | Formula | C29H31N7O |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 152459-95-5 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | STI571, CGP-57148B | Smiles | CC1=C(C=C(C=C1)NC(=O)C2=CC=C(C=C2)CN3CCN(CC3)C)NC4=NC=CC(=N4)C5=CN=CC=C5 | ||
Solubility
|
In vitro |
DMSO
: 98 mg/mL
(198.54 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki | |
|---|---|
| In vitro |
In vitro assays for inhibition of a panel of tyrosine and serine/threonine protein kinases show that Imatinib inhibits the v-Abl tyrosine kinase and PDGFR potently with an IC50 of 0.6 and 0.1 μM, respectively. [1] This compound inhibits the SLF-dependent activation of wild-type c-kit kinase activity with a IC50 for these effects of approximately 0.1 μM, which is similar to the concentration required for inhibition of PDGFR. [2] It exhibits growth-inhibitory activity on the human bronchial carcinoid cell line NCI-H727 and the human pancreatic carcinoid cell line BON-1 with an IC50 of 32.4 and 32.8 μM, respectively. [3] A recent study shows that this chemical has the potential to exert its antileukemia effects in chronic myelogenous leukemia by down-regulating hERG1 K(+) channels, which are highly expressed in leukemia cells and appear of exceptional importance in favoring leukemogenesis. [4] |
| Kinase Assay |
PDGF receptor kinase activity
|
|
PDGF receptor is immunoprecipitated from BALB/c 3T3 cell extracts with rabbit antiserum to the murine PDGF receptor for 2 hours on ice. Protein A-Sepharose beads are used to collect the antigen-antibody complexes. The immunoprecipitates are washed twice with TNET (50 mM Tris, pH 7.5, 140 mM NaCl, 5 mM EDTA, 1% Triton X-100), once with TNE (50 mM Tris, pH 7.5, 140 mM EDTA), and once with kinase buffer (20 mM Tris, pH 7.5,10 mM MgCl2). After stimulation with PDGF (50 ng/mL) for 10 minutes at 4 °C, different concentrations of this compound are added to the reaction mixture. PDGF receptor kinase activity is determined by incubation with 10 μCi [7-33P]-ATP and l μM ATP for 10 minutes at 4 °C. Immune complexes are separated by SDS-PAGE on 7.5% gels.
|
|
| In vivo |
Imatinib produces a different antitumor effect on three xenografted tumors derived from surgical samples of fresh human small cell lung cancers, with 80%, 40% and 78% growth inhibition of SCLC6, SCLC61 and SCLC108 tumors, respectively, and no significant inhibition of SCLC74 growth. [5] In high fat fed ApoE(-/-) mice, this compound significantly reduces the high fat-induced lipid staining area by 30%, 27% and 35% compared to high-fat diet untreated controls when dosed by gavage at 10, 20 and 40 mg/kg, respectively, and suppresses carotid artery lipid accumulation. [6] |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | p-STAT3 / STAT3 / p-STAT5 / STAT5 p-ZAP70 / ZAP70 / p-LAT / LAT p-STAT3 / STAT3 / p-STAT5 / STAT5 p-ZAP70 / ZAP70 / p-LAT / LAT |
|
18981115 |
| Immunofluorescence | RelB Cortactin / F-actin RelB Cortactin / F-actin |
|
20371728 |
| Growth inhibition assay | Cell viability |
|
28435223 |
Clinical Trial Information
(data from https://clinicaltrials.gov, updated on 2024-05-22)
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT05493215 | Recruiting | Gastrointestinal Stromal Tumors |
Reema A. Patel|University of Kentucky |
March 26 2024 | Phase 2 |
| NCT06090669 | Recruiting | Inherited Bone Marrow Failure Syndrome|Familial Platelet Disorder With Predisposition to Myeloid Malignancies |
National Cancer Institute (NCI)|National Institutes of Health Clinical Center (CC) |
December 19 2023 | Phase 1 |
| NCT05413915 | Recruiting | Chronic Myeloid Leukemia|Chronic Myeloid Leukemia BCR/ABL-Positive|Chronic Myeloid Leukemia in Remission |
Sarit Assouline|Novartis|Sir Mortimer B. Davis - Jewish General Hospital |
June 19 2023 | Phase 3 |
| NCT05816785 | Recruiting | Head and Neck Cancer|Squamous Cell Carcinoma of Head and Neck |
University of Wisconsin Madison|National Cancer Institute (NCI) |
April 18 2023 | Early Phase 1 |
| NCT05623774 | Recruiting | CML |
Inhibikase Therapeutics Inc. |
December 16 2022 | Phase 1 |
| NCT05282108 | Recruiting | Chronic Myeloid Leukemia |
Hikma Pharmaceuticals LLC |
July 20 2022 | -- |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
Frequently Asked Questions
Question 1:
Could you please advise whether it is a clear solution for this compound dissolved in vehicle 2% DMSO+30% PEG 300+2% Tween 80+ddH2O?
Answer:
For S2475 (STI571), it is soluble in 2% DMSO+30% PEG 300+2% Tween 80+ddH2O at 2mg/ml. When making the solution, please dissolve this compound in DMSO clearly first. If it dissolves not readily, please sonicate and warm it in water bath at 45-50C. Then add PEG and Tween. After they mixed well, dilute with water.
Question 2:
What is the difference between S2475 (it) and S1026 (its Mesylate)? Are they water soluble?
Answer:
S2475 is free base of this compound while S1026 is a solt form of it. They have exactly the same biological activity but different solubility. S1026 can be dissolved in water, but S2475 is not soluble in water. S2475 can be dissolved in DMSO at up to 3mg/ml.






































