- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
SNF5/SMARCB1 Antibody [D12C14]
Cat.No.: F4832
Application:
Reactivity:
Usage Information
| Dilution |
|---|
|
| Application |
|---|
| WB, IP, IHC, IF, FCM |
| Reactivity |
|---|
| Mouse, Rat, Human |
| Source |
|---|
| Rabbit Monoclonal Antibody |
| Storage Buffer |
|---|
| PBS, pH 7.2+50% Glycerol+0.05% BSA+0.01% NaN3 |
| Storage (from the date of receipt) |
|---|
| -20°C (avoid freeze-thaw cycles), 2 years |
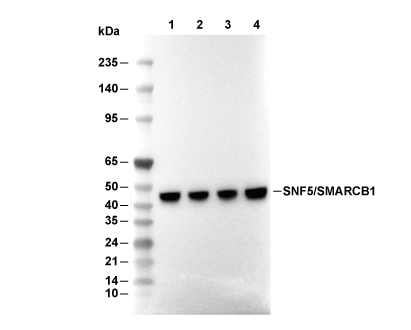
| Predicted MW Observed MW |
|---|
| 44 kDa 44 kDa |
| *Why do the predicted and actual molecular weights differ? The following reasons may explain differences between the predicted and actual protein molecular weight. |
| Positive Control | Rat kidney tissue; Human fetal brain; Human fetal heart; Human kidney tissue; Human fetal spleen; Mouse brain; Mouse spleen; Rat brain; Rat spleen; Mouse kidney tissue; Jurkat cells; HeLa cells; NIH/3T3 cells; PC-12 cells |
|---|---|
| Negative Control | G-401 cells |
Biological Description
| Specificity |
|---|
| SNF5/SMARCB1 Antibody [D12C14] detects endogenous levels of total SNF5/SMARCB1 protein. |
| Clone |
|---|
| D12C14 |
| Synonym(s) |
|---|
| BAF47; INI1; SNF5L1; SMARCB1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1; BRG1-associated factor 47; Integrase interactor 1 protein; SNF5 homolog; Hsnf5 |
| Background |
|---|
| SNF5/SMARCB1 is a core subunit of the mammalian SWI/SNF (BAF) chromatin remodeling complex required for ATP-dependent nucleosome sliding. It maintains lineage fidelity, embryonic stem cell pluripotency, and tumor suppression by facilitating BRG1/BRM ATPase activity at enhancers and promoters. The protein features imperfect repeat motifs in its N-terminal region that enable direct binding to transcription factor DNA-binding surfaces such as MYC:MAX, and contains conserved interfaces that stabilize canonical BAF assembly with ARID1A/B and SS18. SNF5 recruits BAF complexes to differentiation-specific enhancers for H3K27ac deposition and RNA Pol II pause release, and can evict oncogenic drivers like MYC from TSS-proximal sites through competitive DNA binding, independent of chromatin remodeling. Loss of SNF5 destabilizes canonical complexes, leading to residual ncBAF variants that retain super-enhancer function (such as SOX2 regulation in neural tumors) but lose lineage-specific targeting, resulting in enhancer hijacking, MYC de-repression, and lineage infidelity. Biallelic inactivation is found in the majority of malignant rhabdoid tumors (MRTs) and atypical teratoid/rhabdoid tumors (ATRTs), while heterozygous germline mutations increase susceptibility to these pediatric sarcomas via haploinsufficiency. Somatic loss in other cancers, such as renal and medulloblastoma, exploits residual BAF for tumor maintenance. |
| References |
|---|
|
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.