- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
HtrA2/Omi Antibody [H8H4]
Cat.No.: F4813
Application:
Reactivity:
Usage Information
| Dilution |
|---|
|
| Application |
|---|
| WB, IHC, FCM |
| Reactivity |
|---|
| Human |
| Source |
|---|
| Rabbit Monoclonal Antibody |
| Storage Buffer |
|---|
| PBS, pH 7.2+50% Glycerol+0.05% BSA+0.01% NaN3 |
| Storage (from the date of receipt) |
|---|
| -20°C (avoid freeze-thaw cycles), 2 years |
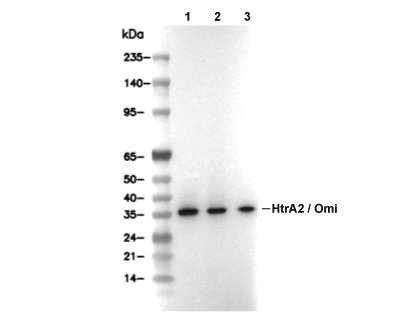
| Predicted MW Observed MW |
|---|
| 49 kDa 38 kDa,43 kDa, 37 kDa |
| *Why do the predicted and actual molecular weights differ? The following reasons may explain differences between the predicted and actual protein molecular weight. |
| Positive Control | Human colon; 293T cells; HeLa cells; Jurkat cells; Human Isolated mitochondria from NB4 cell ine |
|---|---|
| Negative Control |
Biological Description
| Specificity |
|---|
| HtrA2/Omi Antibody [H8H4] detects endogenous levels of total HtrA2/Omi protein. |
| Clone |
|---|
| H8H4 |
| Synonym(s) |
|---|
| OMI; PRSS25; HTRA2; High temperature requirement protein A2; Omi stress-regulated endoprotease; Serine protease 25; Serine proteinase OMI; HtrA2 |
| Background |
|---|
| HtrA2/Omi is a mitochondrial serine protease, homologous to bacterial DegP, that plays a crucial role in protein quality control and apoptosis. It matures from a zymogen to an active form that exposes an N-terminal AVPS motif, which mimics the IAP-binding motif of Smac/DIABLO to interact with inhibitor of apoptosis proteins (IAPs) such as XIAP and cIAP1 via their BIR domains, thereby relieving caspase-3/7/9 inhibition. HtrA2/Omi assembles into a trimeric pyramid with each monomer containing a protease domain characterized by a catalytic triad (Ser-His-Asp), embedded within a framework of α-helices and β-strands, along with regulatory L1, L3, and LD loops and an L2 specificity pocket. The base of the pyramid is formed by a trimeric PDZ domain, which recognizes hydrophobic C-terminal or internal substrate motifs via a carboxylate-binding loop and a defined binding pocket. Interdomain crosstalk between the PDZ and protease domains is critical for activation, as PDZ ligand docking displaces the L3 loop from the buried active site, realigning the catalytic residues for efficient substrate cleavage. Trimerization, stabilized by an N-terminal hydrophobic motif, is essential for allosteric activation and structural integrity. Within mitochondria, HtrA2/Omi degrades misfolded intermembrane space proteins, such as OPA1 fragments and tau aggregates, thus preventing mitochondrial cristae collapse and excessive ROS production. Upon cellular stress, HtrA2/Omi is released into the cytosol, where it amplifies apoptosis by antagonizing IAPs and degrading anti-apoptotic proteins. Loss-of-function mutations that impair PDZ-mediated activation result in neurodegeneration and Parkinson’s disease-like phenotypes due to defective protein quality control, while overexpression in cancer cells promotes metastasis through stabilization of oncogenic pathways such as Notch and β-catenin, and is associated with chemoresistance. |
| References |
|---|
|
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.