- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
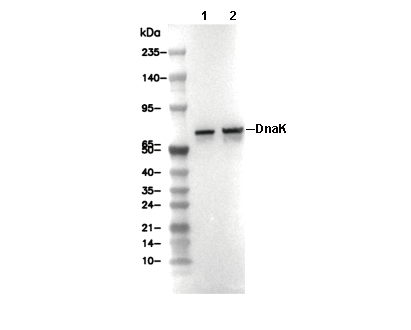
DnaK Antibody [E21A24]
Cat.No.: F2197
Application:
Reactivity:
Usage Information
| Dilution |
|---|
|
| Application |
|---|
| WB |
| Reactivity |
|---|
| Escherichia coli |
| Source |
|---|
| Mouse Monoclonal Antibody |
| Storage Buffer |
|---|
| PBS, pH 7.2+50% Glycerol+0.05% BSA+0.01% NaN3 |
| Storage (from the date of receipt) |
|---|
| -20°C (avoid freeze-thaw cycles), 2 years |
| Predicted MW Observed MW |
|---|
| 70 kDa 70 kDa |
| *Why do the predicted and actual molecular weights differ? The following reasons may explain differences between the predicted and actual protein molecular weight. |
Exprimental Methods
| WB |
|---|
Experimental Protocol:
Sample preparation
1. Tissue: Lyse the tissue sample by adding an appropriate volume of ice-cold RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail),and homogenize the tissue at a low temperature. 2. Adherent cell: Aspirate the culture medium and wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 3. Suspension cell: Transfer the culture medium to a pre-cooled centrifuge tube. Centrifuge and aspirate the supernatant. Wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 4. Place the lysate into a pre-cooled microcentrifuge tube. Centrifuge at 4°C for 15 min. Collect the supernatant;
5. Remove a small volume of lysate to determine the protein concentration;
6. Combine the lysate with protein loading buffer. Boil 20 µL sample under 95-100°C for 5 min. Centrifuge for 5 min after cool down on ice.
Electrophoretic separation
1. According to the concentration of extracted protein, load appropriate amount of protein sample and marker onto SDS-PAGE gels for electrophoresis. Recommended separating gel (lower gel) concentration: 10%. Reference Table for Selecting SDS-PAGE Separation Gel Concentrations 2. Power up 80V for 30 minutes. Then the power supply is adjusted (110 V~150 V), the Marker is observed, and the electrophoresis can be stopped when the indicator band of the predyed protein Marker where the protein is located is properly separated. (Note that the current should not be too large when electrophoresis, too large current (more than 150 mA) will cause the temperature to rise, affecting the result of running glue. If high currents cannot be avoided, an ice bath can be used to cool the bath.)
Transfer membrane
1. Take out the converter, soak the clip and consumables in the pre-cooled converter;
2. Activate PVDF membrane with methanol for 1 min and rinse with transfer buffer;
3. Install it in the order of "black edge of clip - sponge - filter paper - filter paper - glue -PVDF membrane - filter paper - filter paper - sponge - white edge of clip"; 4. The protein was electrotransferred to PVDF membrane. ( 0.45 µm PVDF membrane is recommended ) Reference Table for Selecting PVDF Membrane Pore Size Specifications Recommended conditions for wet transfer: 200 mA, 120 min. ( Note that the transfer conditions can be adjusted according to the protein size. For high-molecular-weight proteins, a higher current and longer transfer time are recommended. However, ensure that the transfer tank remains at a low temperature to prevent gel melting.)
Block
1. After electrotransfer, wash the film with TBST at room temperature for 5 minutes;
2. Incubate the film in the blocking solution for 1 hour at room temperature;
3. Wash the film with TBST for 3 times, 5 minutes each time.
Antibody incubation
1. Use 5% skim milk powder to prepare the primary antibody working liquid (recommended dilution ratio for primary antibody 1:1000), gently shake and incubate with the film at 4°C overnight; 2. Wash the film with TBST 3 times, 5 minutes each time;
3. Add the secondary antibody to the blocking solution and incubate with the film gently at room temperature for 1 hour;
4. After incubation, wash the film with TBST 3 times for 5 minutes each time.
Antibody staining
1. Add the prepared ECL luminescent substrate (or select other color developing substrate according to the second antibody) and mix evenly;
2. Incubate with the film for 1 minute, remove excess substrate (keep the film moist), wrap with plastic film, and expose in the imaging system. |
Biological Description
| Specificity |
|---|
| DnaK Antibody [E21A24] detects endogenous levels of total DnaK protein. |
| Subcellular Location |
|---|
| Cell inner membrane, Cell membrane, Cytoplasm, Membrane |
| Uniprot ID |
|---|
| P0A6Y8 |
| Clone |
|---|
| E21A24 |
| Synonym(s) |
|---|
| groP; grpF; seg; b0014; JW0013; dnaK; Chaperone protein DnaK; HSP70; Heat shock 70 kDa protein; Heat shock protein 70 |
| Background |
|---|
| DnaK is a highly conserved bacterial heat shock protein belonging to the HSP70 family, primarily studied in Escherichia coli, where it functions as a molecular chaperone crucial for protein folding, refolding of denatured proteins, and protein homeostasis under stress conditions. DnaK consists of two main domains: a 44-kDa N-terminal nucleotide-binding domain (NBD) responsible for ATP binding and hydrolysis, and a 25-kDa C-terminal substrate-binding domain (SBD), which includes a β-sandwich subdomain that binds unfolded protein substrates via exposed hydrophobic patches. DnaK operates through an ATP-dependent cycle where ATP binding induces an open conformation allowing substrate release, while ATP hydrolysis triggers a closed conformation leading to high-affinity substrate binding. DnaK forms transient dimers in its ATP-bound state, which enhances its interaction with the co-chaperone DnaJ (Hsp40), facilitating efficient substrate recognition and delivery. The co-chaperone GrpE acts as a nucleotide exchange factor, accelerating ADP release and resetting DnaK for another cycle. DnaK helps prevent protein aggregation during cellular stress and participates in critical processes such as DNA replication, mediated by interactions with the DnaJ co-chaperone and other partners. This chaperone network is essential for survival under heat shock and other proteotoxic stresses, maintaining protein quality control and cellular homeostasis. |
| References |
|---|
|
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.