research use only
ABCA1 Antibody [D20K13]
Cat.No.: F1630
Application:
Reactivity:
Experiment Essentials
WB
Recommended SDS-PAGE separating gel concentration: 5%.
Recommended wet transfer conditions: 250 mA, 180 min.
Recommended WB dilution ratio: 1:200
Recommended SDS-PAGE separating gel concentration: 5%.
Recommended wet transfer conditions: 250 mA, 180 min.
Recommended WB dilution ratio: 1:200
Usage Information
| Dilution |
|---|
|
| Application |
|---|
| WB, IHC, FCM |
| Reactivity |
|---|
| Mouse, Human |
| Source |
|---|
| Mouse Monoclonal Antibody |
| Storage Buffer |
|---|
| PBS, pH 7.2+50% Glycerol+0.05% BSA+0.01% NaN3 |
| Storage (from the date of receipt) |
|---|
| -20°C (avoid freeze-thaw cycles), 2 years |
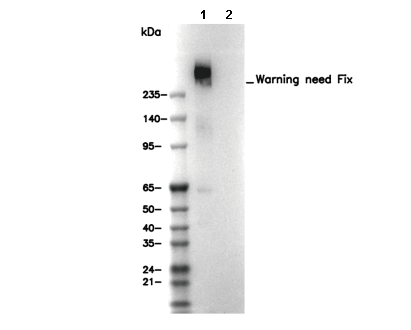
| Predicted MW Observed MW |
|---|
| 254 kDa 254 kDa |
| *Why do the predicted and actual molecular weights differ? The following reasons may explain differences between the predicted and actual protein molecular weight. |
| Positive Control | Pancreas tissue; Mouse liver tissue; HepG2 cells; Human fibroblast cells (cholesterol treated) |
|---|---|
| Negative Control | Human fibroblast cells |
Exprimental Methods
| WB |
|---|
Experimental Protocol:
Sample preparation
1. Tissue: Lyse the tissue sample by adding an appropriate volume of ice-cold RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail),and homogenize the tissue at a low temperature. 2. Adherent cell: Aspirate the culture medium and wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 3. Suspension cell: Transfer the culture medium to a pre-cooled centrifuge tube. Centrifuge and aspirate the supernatant. Wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 4. Place the lysate into a pre-cooled microcentrifuge tube. Centrifuge at 4°C for 15 min. Collect the supernatant;
5. Remove a small volume of lysate to determine the protein concentration;
6. Combine the lysate with protein loading buffer. Boil 20 µL sample under 95-100°C for 5 min. Centrifuge for 5 min after cool down on ice.
Electrophoretic separation
1. According to the concentration of extracted protein, load appropriate amount of protein sample and marker onto SDS-PAGE gels for electrophoresis. Recommended separating gel (lower gel) concentration: 5%. Reference Table for Selecting SDS-PAGE Separation Gel Concentrations 2. Power up 80V for 30 minutes. Then the power supply is adjusted (110 V~150 V), the Marker is observed, and the electrophoresis can be stopped when the indicator band of the predyed protein Marker where the protein is located is properly separated. (Note that the current should not be too large when electrophoresis, too large current (more than 150 mA) will cause the temperature to rise, affecting the result of running glue. If high currents cannot be avoided, an ice bath can be used to cool the bath.)
Transfer membrane
1. Take out the converter, soak the clip and consumables in the pre-cooled converter;
2. Activate PVDF membrane with methanol for 1 min and rinse with transfer buffer;
3. Install it in the order of "black edge of clip - sponge - filter paper - filter paper - glue -PVDF membrane - filter paper - filter paper - sponge - white edge of clip"; 4. The protein was electrotransferred to PVDF membrane. ( 0.45 µm PVDF membrane is recommended ) Reference Table for Selecting PVDF Membrane Pore Size Specifications Recommended conditions for wet transfer: 250 mA, 180 min. ( Note that the transfer conditions can be adjusted according to the protein size. For high-molecular-weight proteins, a higher current and longer transfer time are recommended. However, ensure that the transfer tank remains at a low temperature to prevent gel melting.)
Block
1. After electrotransfer, wash the film with TBST at room temperature for 5 minutes;
2. Incubate the film in the blocking solution for 1 hour at room temperature;
3. Wash the film with TBST for 3 times, 5 minutes each time.
Antibody incubation
1. Use 5% skim milk powder to prepare the primary antibody working liquid (recommended dilution ratio for primary antibody 1:200), gently shake and incubate with the film at 4°C overnight; 2. Wash the film with TBST 3 times, 5 minutes each time;
3. Add the secondary antibody to the blocking solution and incubate with the film gently at room temperature for 1 hour;
4. After incubation, wash the film with TBST 3 times for 5 minutes each time.
Antibody staining
1. Add the prepared ECL luminescent substrate (or select other color developing substrate according to the second antibody) and mix evenly;
2. Incubate with the film for 1 minute, remove excess substrate (keep the film moist), wrap with plastic film, and expose in the imaging system. |
| IHC |
|---|
Experimental Protocol:
Deparaffinization/Rehydration
1. Deparaffinize/hydrate sections:
2. Incubate sections in three washes of xylene for 5 min each.
3. Incubate sections in two washes of 100% ethanol for 10 min each.
4. Incubate sections in two washes of 95% ethanol for 10 min each.
5. Wash sections two times in dH2O for 5 min each.
6.Antigen retrieval: For Citrate: Heat slides in a microwave submersed in 1X citrate unmasking solution until boiling is initiated; continue with 10 min at a sub-boiling temperature (95°-98°C). Cool slides on bench top for 30 min.
Staining
1. Wash sections in dH2O three times for 5 min each.
2. Incubate sections in 3% hydrogen peroxide for 10 min.
3. Wash sections in dH2O two times for 5 min each.
4. Wash sections in wash buffer for 5 min.
5. Block each section with 100–400 µl of blocking solution for 1 hr at room temperature.
6. Remove blocking solution and add 100–400 µl primary antibody diluent in to each section. Incubate overnight at 4°C.
7. Remove antibody solution and wash sections with wash buffer three times for 5 min each.
8. Cover section with 1–3 drops HRPas needed. Incubate in a humidified chamber for 30 min at room temperature.
9. Wash sections three times with wash buffer for 5 min each.
10. Add DAB Chromogen Concentrate to DAB Diluent and mix well before use.
11. Apply 100–400 µl DAB to each section and monitor closely. 1–10 min generally provides an acceptable staining intensity.
12. Immerse slides in dH2O.
13. If desired, counterstain sections with hematoxylin.
14. Wash sections in dH2O two times for 5 min each.
15. Dehydrate sections: Incubate sections in 95% ethanol two times for 10 sec each; Repeat in 100% ethanol, incubating sections two times for 10 sec each; Repeat in xylene, incubating sections two times for 10 sec each.
16. Mount sections with coverslips and mounting medium.
|
Biological Description
| Specificity |
|---|
| ABCA1 Antibody [D20K13] detects endogenous levels of total ABCA1 protein. |
| Subcellular Location |
|---|
| Cell membrane, Endosome, Membrane |
| Uniprot ID |
|---|
| O95477 |
| Clone |
|---|
| D20K13 |
| Synonym(s) |
|---|
| ABC1; CERP; ABCA1; Phospholipid-transporting ATPase ABCA1; ATP-binding cassette sub-family A member 1; ATP-binding cassette transporter 1; Cholesterol efflux regulatory protein; ABC-1; ATP-binding cassette 1 |
| Background |
|---|
| ATP binding cassette transporter A1 (ABCA1) is a large membrane transporter of the ABC family that uses ATP hydrolysis to export cellular cholesterol and phospholipids to lipid poor apolipoproteins, especially apoA I, thereby initiating nascent HDL formation and driving the first, rate limiting step of reverse cholesterol transport from peripheral tissues to the liver. ABCA1 is a protein comprising two six helix transmembrane domains that create a lipid translocation pathway, two cytosolic nucleotide binding domains with Walker A/B and LSGGQ signature motifs for ATP binding and hydrolysis, and two large, glycosylated extracellular domains unique to the ABCA subfamily that form a hydrophobic surface or tunnel for apoA I docking and lipid loading, with additional regulatory features such as a PEST sequence controlling proteolytic turnover and palmitoylated cysteines important for plasma membrane targeting. Cholesterol loading activates LXR/RXR, which upregulates ABCA1; at the plasma membrane and endosomes, ABCA1 undergoes ATP driven conformational cycling to mobilize phospholipids and free cholesterol from the inner leaflet and intracellular pools to the cell surface, where apoA I binding both stabilizes ABCA1 and captures these lipids to form discoidal nascent HDL, while concomitantly disrupting cholesterol rich lipid rafts and triggering JAK2/STAT3 and other signaling pathways that dampen inflammation and promote further efflux in macrophages. Through these transport and signaling functions, ABCA1 is essential for preventing foam cell formation and atherosclerosis and also contributes to β cell function, insulin secretion, and brain apoE lipidation and amyloid β handling; loss of function mutations in ABCA1 cause Tangier disease and familial HDL deficiency with near absent HDL and cholesterol ester accumulation, whereas more subtle defects in ABCA1 expression or activity increase cardiovascular and metabolic risk and may modulate susceptibility to late onset Alzheimer’s disease. |
| References |
|---|
|
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.