- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at [email protected] to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly FLAG Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
UNC1999
UNC1999 is a potent, orally bioavailable and selective inhibitor of EZH2 and EZH1 with IC50 of 2 nM and 45 nM in cell-free assays, respectively, showing >1000-fold selectivity over a broad range of epigenetic and non-epigenetic targets. UNC1999 is a potent autophagy inducer. UNC1999 specifically suppresses H3K27me3/2 and induces a range of anti-leukemia effects including anti-proliferation, differentiation, and apoptosis.
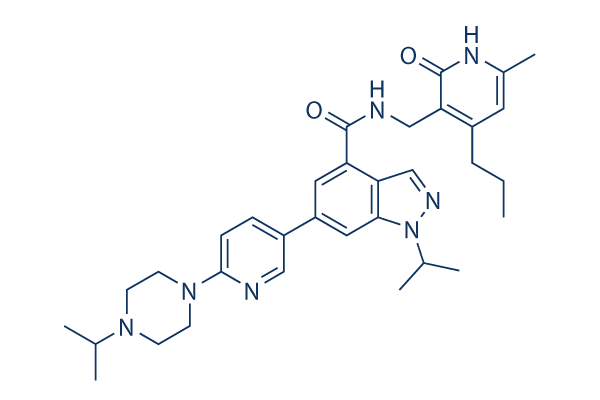
UNC1999 Chemical Structure
CAS: 1431612-23-5
Selleck's UNC1999 has been cited by 25 Publications
2 Customer Reviews
Purity & Quality Control
Batch:
Purity:
99.99%
99.99
UNC1999 Related Products
Signaling Pathway
Choose Selective Histone Methyltransferase Inhibitors
Cell Data
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| human MCF10A cells | Function assay | 72 h | Inhibition of EZH2 in human MCF10A cells assessed as reduction of H3K27me3 level after 72 hrs by Western blot analysis, IC50=0.124 μM | 25406853 | ||
| human MCF10A cells | Cytotoxic assay | Cytotoxicity against human MCF10A cells assessed as cell viability by Alamar Blue assay, EC50=19.2 μM | 25406853 | |||
| Click to View More Cell Line Experimental Data | ||||||
Biological Activity
| Description | UNC1999 is a potent, orally bioavailable and selective inhibitor of EZH2 and EZH1 with IC50 of 2 nM and 45 nM in cell-free assays, respectively, showing >1000-fold selectivity over a broad range of epigenetic and non-epigenetic targets. UNC1999 is a potent autophagy inducer. UNC1999 specifically suppresses H3K27me3/2 and induces a range of anti-leukemia effects including anti-proliferation, differentiation, and apoptosis. | ||||
|---|---|---|---|---|---|
| Features | The first orally bioavailable inhibitor against wild-type and mutant EZH2 as well as EZH1. | ||||
| Targets |
|
| In vitro | ||||
| In vitro | UNC1999 is highly potent for both EZH2 Y641N and EZH2 Y641F mutants in vitro. UNC1999 causes concentration-dependent reductions of H3K27me3 in MCF10A cells with IC50 of 124 nM , while shows low cellular toxicity. UNC1999 displays potent, concentration-dependent inhibition of cell proliferation with EC50 of 633 nM in a DLBCL cell line harboring the EZH2Y641N mutant. In addition, biotinylated UNC1999 enriches EZH2 from HEK293T cell lysates, and thus may be used in chemoproteomics studies. [1] | |||
|---|---|---|---|---|
| Kinase Assay | Scintillation Proximity Assay | |||
| Methyltransferase activity assays are performed by monitoring the incorporation of tritiumlabeled methyl group from S-adenosylmethionine (3H-SAM) to biotinylated peptide substrates using Scintillation Proximity Assay (SPA) for PRC2-EZH2 trimeric complex (EZH2:EED:SUZ12), PRC2-EZH1 pentameric complex (EZH1:EED:SUZ12:RBBP4:AEBP2), SETD7, G9a, GLP, SETDB1, SETD8, SUV420H1, SUV420H2, SUV39H2, MLL1 tetrameric complex (MLL:WDR5:RbBP5:ASH2L), PRMT1, PRMT3, PRMT5-MEP50 complex and SMYD2. The reaction buffer for SMYD2 and SMYD3 is 50 mM Tris pH 9.0, 5 mM DTT, 0.01% TritonX-100; for G9a, GLP and SUV39H2 is 25 mM potassium phosphate pH 8.0, 1 mM EDTA, 2 mM MgCl2 and 0.01% Triton X-100; and for other HMTs 20 mM Tris pH 8.0, 5 mM DTT, 0.01% TritonX-100. To stop the enzymatic reactions, 10 μL of 7.5 M guanidine hydrochloride is added, followed by 180 μL of buffer, mixed and transferred to a 384-well FlashPlate. After mixing, the reaction mixtures are incubated and the CPM counts are measured using Topcount plate reader. The CPM counts in the absence of compound for each data set are defined as 100% activity. In the absence of the enzyme, the CPM counts in each data set are defined as background (0%). IC50 values are determined using compound concentrations ranging from 100 nM to 100 μM. The IC50 values are determined using SigmaPlot software. EZH2-Y641F assays are performed using 30 nM of enzyme in 20 mM Tris pH 8, 5 mM DTT, 0.01% Triton X-100, 5 μM SAM and 1 μM of H3 (1-24) peptide (same as for the wild-type PRC2-EZH2 complex). For DNMT1, the assay is performed using hemimethylated dsDNA as a substrate. The dsDNA substrate is prepared by annealing two complementary strands (biotintlated forward strand: BGAGCCCGTAAGCCCGTTCAGGTCG and reverse strand: CGACCTGAACGGGCTTACGGGCTC), synthesized by Eurofins MWG Operon. Reaction buffer is 20 mM Tris-HCl, pH 8.0, 5mM DTT, 0.01% Triton X-100. Methyltransferase activity assays for DOT1L is performed using Filter-plates. Reaction mixtures in 20 mM Tris-HCl, pH 8.0, 5 mM DTT, 2 mM MgCl2 and 0.01% Triton X-100 are incubated at room temperature for 1h, 100 μL 10% TCA is added, mixed and transferred to filter-plate. Plates are centrifuged at 2000 rpm for 2 min followed by 2 additional 10% TCA wash and one ethanol wash (180 μL) followed by centrifugation. Plates are dried and 100 μL MicroO is added and centrifuged. 70 μL MicroO is added and CPM are measured using Topcount plate reader. | ||||
| Cell Research | Cell lines | DLBCL cell line harboring the EZH2Y641N mutant | ||
| Concentrations | ~5 μM | |||
| Incubation Time | 8 days | |||
| Method | DB cells, a diffuse-large B-cell lymphoma cell line harboring the EZH2 Y641N mutation, are obtained from ATCC and cultured in RPMI 1640 supplemented with 10% fetal bovine serum, antibiotics, and various concentrations of compounds (DMSO control, UNC1999, or UNC2400). The medium containing the test compound or control is refreshed every 3 days. The numbers of viable cells from at least three independent experiments are measured using TC20 automated cell counter system. Total histones are prepared from cell nuclei using an acidic extraction protocol. About 1 μg of total histones is separated using 15% SDS-PAGE, transferred to PVDF membranes, and probed with histone antibodies. Antibodies used in this study are those against EZH2, general H3, and H3K27me3. |
|||
| Experimental Result Images | Methods | Biomarkers | Images | PMID |
| Western blot | H3K27Me3 / Cleaved PARP / LC3B H3K27me2 / H3K27me1 / H3K27ac / H3K4me3 / H3K9me2 / H3K36me2 |
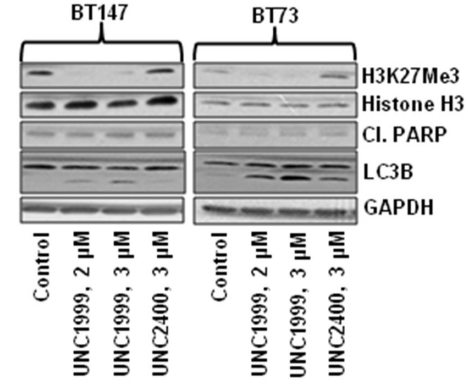
|
27449082 | |
| Immunofluorescence | EZH2 / H3K27me3 NF-κB |
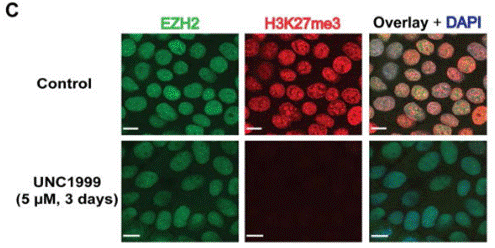
|
23614352 | |
| Growth inhibition assay | Cell viability |
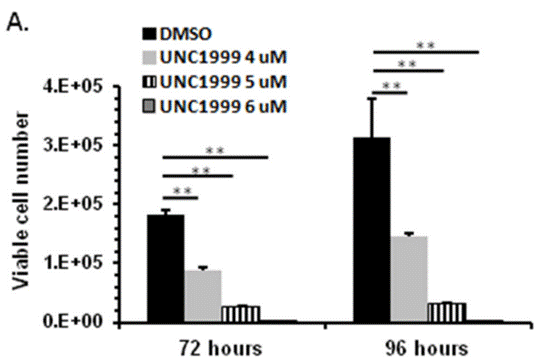
|
27449082 | |
| In Vivo | ||
| In vivo | Treatment of UNC1999 (150 and 50 mg/kg, i.p.) results in the plasma concentrations of UNC1999 above its cellular IC50 over 24 hours in vivo. In addition, UNC1999 is also orally bioavailable in mice, which makes chronic animal studies more practical and convenient. [1] | |
|---|---|---|
| Animal Research | Animal Models | Male Swiss albino mice |
| Dosages | ~150 mg/kg (i.p.), ~50 mg/kg (oral) | |
| Administration | Intraperitoneal administration or oral administration | |
Chemical Information & Solubility
| Molecular Weight | 569.74 | Formula | C33H43N7O2 |
| CAS No. | 1431612-23-5 | SDF | Download UNC1999 SDF |
| Smiles | CCCC1=C(C(=O)NC(=C1)C)CNC(=O)C2=C3C=NN(C3=CC(=C2)C4=CN=C(C=C4)N5CCN(CC5)C(C)C)C(C)C | ||
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 50 mg/mL ( (87.75 mM); Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Ethanol : 50 mg/mL Water : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field
Tags: buy UNC1999 | UNC1999 supplier | purchase UNC1999 | UNC1999 cost | UNC1999 manufacturer | order UNC1999 | UNC1999 distributor