- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at [email protected] to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly FLAG Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
Niclosamide
Synonyms: BAY2353, Niclocide, NSC 178296
Niclosamide can inhibit DNA replication and inhibit STAT3 with IC50 of 0.7 μM in a cell-free assay. Niclosamide selectively inhibited the phosphorylation of STAT3 and had no obvious inhibition against the activation of other homologues (e.g., STAT1 and STAT5).
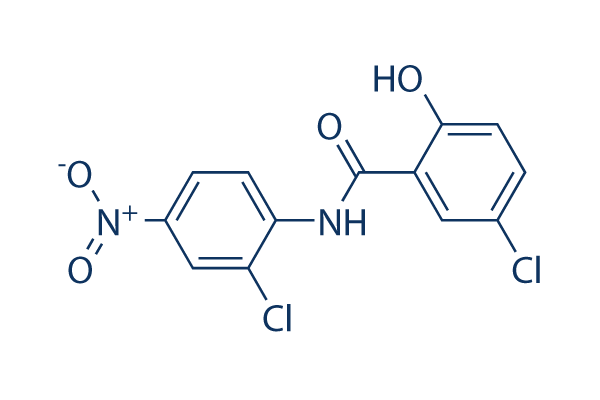
Niclosamide Chemical Structure
CAS: 50-65-7
Selleck's Niclosamide has been cited by 46 publications
Purity & Quality Control
Batch:
Purity:
99.98%
99.98
Niclosamide Related Products
| Related Targets | STAT1 STAT3 STAT5 STAT6 | Click to Expand |
|---|---|---|
| Related Compound Libraries | Kinase Inhibitor Library FDA-approved Drug Library Natural Product Library Tyrosine Kinase Inhibitor Library JAK/STAT compound library | Click to Expand |
Signaling Pathway
Choose Selective STAT Inhibitors
Cell Data
| Cell Lines | Assay Type | Concentration | Incubation Time | Formulation | Activity Description | PMID |
|---|---|---|---|---|---|---|
| PC3 | Antiproliferative assay | 120 hrs | Antiproliferative activity against human PC3 cells after 120 hrs by MTT assay, IC50=0.4μM | 16680159 | ||
| A549 | Antiproliferative assay | 120 hrs | Antiproliferative activity against human A549 cells after 120 hrs by MTT assay, IC50=0.4μM | 16680159 | ||
| U87MG | Antiproliferative assay | 120 hrs | Antiproliferative activity against human U87MG cells after 120 hrs by MTT assay, IC50=0.4μM | 16680159 | ||
| LoVo | Antiproliferative assay | 120 hrs | Antiproliferative activity against human LoVo cells after 120 hrs by MTT assay, IC50=0.7μM | 16680159 | ||
| MIAPaCa2 | Antiproliferative assay | 120 hrs | Antiproliferative activity against human MIAPaCa2cells after 120 hrs by MTT assay, IC50=1.1μM | 16680159 | ||
| PC3 | Function assay | 1 hr | Inhibition of mitochondrial membrane potential in human PC3 cells after 1 hr | 16680159 | ||
| HEK293 | Function assay | 1 hr | Inhibition of mitochondrial membrane potential in human HEK293 cells after 1 hr | 16680159 | ||
| neural precursor | Function assay | Inhibition of neurosphere proliferation of mouse neural precursor cells by MTT assay | 17417631 | |||
| Vero E6 | Antiviral assay | Antiviral activity against SARS coronavirus in Vero E6 cells assessed as inhibition of viral replication by ELISA, EC50<0.1μM | 17663539 | |||
| Vero E6 | Cytotoxicity assay | Cytotoxicity against Vero E6 cells by MTT assay, CC50=22.1μM | 17663539 | |||
| RAW264.7 | Cytoprotective assay | 10 uM | Cytoprotective activity against anthrax toxin lethal factor/protective antigen-induced cell death in mouse RAW264.7 cells assessed as cell viability at 10 uM by MTT reduction assay | 19540764 | ||
| CHO | Cytoprotective assay | Cytoprotective activity against anthrax fusion toxin FP59-induced cell death in CHO cells assessed as cell viability by MTT reduction assay | 19540764 | |||
| CHO | Function assay | Inhibition of Bacillus anthracis anthrax protective antigen heptamer pre-pore to pore conversion in CMG2-expressing CHO cells | 19540764 | |||
| Ava5 | Antiviral assay | 3 days | Antiviral activity against HCV genotype 1b infected in Ava5 cells assessed as inhibition of viral replication after 3 days by blot hybridization analysis, EC50=0.16μM | 22059983 | ||
| Ava5 | Antiviral assay | 3 days | Antiviral activity against HCV genotype 1b infected in Ava5 cells assessed as inhibition of viral replication after 3 days by blot hybridization analysis, EC90=0.7μM | 22059983 | ||
| Ava5 | Cytotoxicity assay | 3 days | Cytotoxicity against human Ava5 cells after 3 days by neutral red dye assay, CC50=10μM | 22059983 | ||
| HFF | Antiapicomplexan assay | 24 hrs | Antiapicomplexan activity against Toxoplasma gondii RH tachyzoites infected in HFF cells assessed as [3H]-uracil incorporation after 24 hrs by scintillation luminescence counter, MIC90=0.2μM | 22970937 | ||
| HFF | Antiapicomplexan assay | 24 hrs | Antiapicomplexan activity against Toxoplasma gondii RH tachyzoites infected in HFF cells assessed as [3H]-uracil incorporation after 24 hrs by scintillation luminescence counter, MIC50=0.2μM | 22970937 | ||
| MDA-MB-231 | Cytotoxicity assay | CYtotoxicity against ER-negative human MDA-MB-231 cells by MTS assay, IC50=0.79μM | 23416191 | |||
| MCF7 | Cytotoxicity assay | CYtotoxicity against ER-positive human MCF7 cells by MTS assay, IC50=1.06μM | 23416191 | |||
| AsPC1 | Cytotoxicity assay | CYtotoxicity against human AsPC1 cells by MTS assay, IC50=1.47μM | 23416191 | |||
| PANC1 | Cytotoxicity assay | CYtotoxicity against human PANC1 cells by MTS assay, IC50=1.73μM | 23416191 | |||
| HEK293 | Function assay | 8 hrs | Inhibition of Wnt3A/beta-casein signaling in HEK293 cells after 8 hrs by TOPflash reporter assay, IC50=0.4μM | 23453073 | ||
| U2OS | Function assay | 12.5 uM | 6 hrs | Induction of internalization of Frizzled1-GFP (unknown origin) expressed in human U2OS cells at 12.5 uM after 6 hrs by confocal microscopy | 23453073 | |
| MDA-MB-231 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human ER negative MDA-MB-231 cells after 72 hrs by MTS assay, IC50=0.79μM | 23459613 | ||
| MCF7 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human ER positive MCF7 cells after 72 hrs by MTS assay, IC50=1.06μM | 23459613 | ||
| AsPC1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human AsPC1 cells after 72 hrs by MTS assay, IC50=1.47μM | 23459613 | ||
| PANC1 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human PANC1 cells after 72 hrs by MTS assay, IC50=1.73μM | 23459613 | ||
| MDA-MB-231 | Antitumor assay | 75 mg/kg | 5 days | Antitumor activity against human ER negative MDA-MB-231 cells xenografted in nude mouse assessed as inhibition of tumor growth at 75 mg/kg, po qd for 5 days | 23459613 | |
| MDA-MB-231 | Antitumor assay | 12.5 mg/kg | 5 days | Antitumor activity against human ER negative MDA-MB-231 cells xenografted in nude mouse assessed as inhibition of tumor growth at 12.5 mg/kg, ip qd for 5 days | 23459613 | |
| MDA-MB-231 | Antiproliferative assay | 10 to 20 uM | 24 hrs | Antiproliferative activity against human ER negative MDA-MB-231 cells at 10 to 20 uM after 24 hrs by MTT assay | 23459613 | |
| MDA-MB-231 | Function assay | 24 hrs | Inhibition of STAT3 promoter activity in human ER negative MDA-MB-231 cells after 24 hrs by dual luciferase reporter assay | 23459613 | ||
| MDA-MB-231 | Function assay | 20 uM | 24 hrs | Inhibition of STAT3 promoter activity in human ER negative MDA-MB-231 cells at 20 uM after 24 hrs by dual luciferase reporter assay | 23459613 | |
| MDA-MB-231 | Function assay | 10 uM | 24 hrs | Inhibition of STAT3 promoter activity in human ER negative MDA-MB-231 cells at 10 uM after 24 hrs by dual luciferase reporter assay | 23459613 | |
| MDA-MB-231 | Function assay | 1 to 10 uM | 24 hrs | Inhibition of STAT3 in human ER negative MDA-MB-231 cells assessed as reduction of total STAT3 level at 1 to 10 uM after 24 hrs by Western blot analysis | 23459613 | |
| MDA-MB-231 | Function assay | 1 to 10 uM | 48 hrs | Induction of morphological changes in human ER negative MDA-MB-231 cells at 1 to 10 uM after 48 hrs by light microscopic analysis | 23459613 | |
| MDA-MB-231 | Function assay | 1 to 10 uM | 48 hrs | Induction of apoptosis in human ER negative MDA-MB-231 cells at 1 to 10 uM after 48 hrs by light microscopic analysis | 23459613 | |
| MDA-MB-231 | Antiproliferative assay | 1 to 10 uM | 48 hrs | Antiproliferative activity against human ER negative MDA-MB-231 cells at 1 to 10 uM after 48 hrs by light microscopic analysis | 23459613 | |
| MDA-MB-231 | Function assay | 1 to 10 uM | 24 hrs | Inhibition of STAT3 in human ER negative MDA-MB-231 cells assessed as reduction of phosphorylated STAT3 at Tyr-705 level at 1 to 10 uM after 24 hrs by Western blot analysis | 23459613 | |
| HeLa | Function assay | 24 hrs | Inhibition of STAT3 in human HeLa cells after 24 hrs by luciferase reporter gene assay, IC50=0.25μM | 24900231 | ||
| DU145 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human DU145 cells after 72 hrs by MTT assay, IC50=0.7μM | 24900231 | ||
| DU145 | Growth inhibition assay | 11 to 12 days | Growth inhibition of human DU145 cells assessed as inhibition of colony formation after 11 to 12 days by crystal violet staining based microscopic analysis, IC50=0.7μM | 24900231 | ||
| HeLa | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HeLa cells after 72 hrs by MTT assay, IC50=1.4μM | 24900231 | ||
| A549 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A549 cells after 72 hrs by MTT assay, IC50=3μM | 24900231 | ||
| HT-29 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HT-29 cells after 72 hrs by MTT assay, IC50=7.2μM | 24900231 | ||
| A431 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human A431 cells after 72 hrs by MTT assay, IC50=8.8μM | 24900231 | ||
| PC3 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human PC3 cells after 72 hrs by MTT assay, IC50=11.7μM | 24900231 | ||
| HeLa | Function assay | 5 uM | 24 hrs | Inhibition of STAT3 in human HeLa cells at 5 uM after 24 hrs by luciferase reporter gene assay | 24900231 | |
| DU145 | Function assay | 1 uM | 2 hrs | Inhibition of STAT3 nuclear translocation in EGF stimulated human DU145 cells at 1 uM administered before 100 ng/ml EGF stimulation measured after 2 hrs by confocal laser microscopy | 24900231 | |
| DU145 | Function assay | 05 to 10 uM | 2 hrs | Inhibition of STAT3 interaction with DNA binding site in human DU145 cells at 05 to 10 uM after 2 hrs by EMSA | 24900231 | |
| human | Function assay | 0.1 to 2 uM | 2 hrs | Decrease in cyclin D1 protein level in human DU145 cells at 0.1 to 2 uM after 2 hrs by Western blot analysis | 24900231 | |
| human | Function assay | 0.1 to 2 uM | 2 hrs | Decrease in c-Myc protein level in human DU145 cells at 0.1 to 2 uM after 2 hrs by Western blot analysis | 24900231 | |
| human | Function assay | 0.1 to 2 uM | 2 hrs | Decrease in Bcl-Xl protein level in human DU145 cells at 0.1 to 2 uM after 2 hrs by Western blot analysis | 24900231 | |
| DU145 | Function assay | 0.1 to 2 uM | 2 hrs | Downregulation of Mcl-1 protein in human DU145 cells at 0.1 to 2 uM after 2 hrs by Western blot analysis | 24900231 | |
| human | Function assay | 2 uM | 2 hrs | Inhibition of STAT3 phosphorylation at Tyr705 in human DU145 cells at 2 uM within 2 hrs by Western blot analysis | 24900231 | |
| breast cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human breast cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| colon cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human colon cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| lung cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human lung cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| prostate cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human prostate cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| ovarian cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human ovarian cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| blood cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human blood cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| pancreatic cancer cells | Cytotoxicity assay | 72 hrs | Cytotoxicity against human pancreatic cancer cells assessed as growth inhibition after 72 hrs by Cell Titer Glo Assay, IC50=0.13μM | 26272032 | ||
| HEK293 | Function assay | 8 hrs | Inhibition of Wnt/beta-catenin in HEK293 cells assessed as inhibition of Wnt3A-stimulated TOPFlash activity after 8 hrs, IC50=0.34μM | 26272032 | ||
| HCT116 | Cytotoxicity assay | 72 hrs | Cytotoxicity against human HCT116 cells assessed as growth inhibition after 72 hrs by MTS assay, IC50=0.45μM | 26272032 | ||
| U2OS | Function assay | 12.5 uM | 6 hrs | Inhibition of Wnt/beta-catenin in human U2OS cells assessed as internalization of frizzled-GFP at 12.5 uM after 6 hrs by confocal microscopic analysis | 26272032 | |
| HCT116 | Function assay | 3 hrs | Effect on ATP homeostasis in human HCT116 cells assessed as reduction in ATP level after 3 hrs in absence of glucose by ATP bioluminescent assay, EC50=0.056μM | 28233680 | ||
| HEK293 | Function assay | 6 hrs | Inhibition of Wnt/beta-catenin signaling (unknown origin) expressed in HEK293 cells assessed as inhibition of Wnt3A-stimulated beta-catenin response transcription after 6 hrs by TOPflash dual luciferase reporter gene assay, IC50=0.12μM | 28233680 | ||
| HCT116 | Function assay | 3 hrs | Effect on ATP homeostasis in human HCT116 cells assessed as reduction in ATP level after 3 hrs in absence of glucose by ATP bioluminescent assay, IC50=0.25μM | 28233680 | ||
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human HCT116 cells assessed as downregulation of cytosolic beta-catenin protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human HCT116 cells assessed as downregulation of cytosolic Axin2 protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human HCT116 cells assessed as downregulation of c-Myc protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human HCT116 cells assessed as downregulation of Cyclin D1 protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human HCT116 cells assessed as downregulation of survivin protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human SW480 cells assessed as downregulation of cytosolic beta-catenin protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human SW480 cells assessed as downregulation of cytosolic Axin2 protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human SW480 cells assessed as downregulation of c-Myc protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human SW480 cells assessed as downregulation of Cyclin D1 protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling in human SW480 cells assessed as downregulation of survivin protein expression at 5 uM after 18 hrs by Western blot method | 28233680 | |
| HCT116 | Function assay | 2 uM | 30 mins | Induction of AMPK phosphorylation at Thr-172 residue in human HCT116 cells at 2 uM after 30 mins in glucose supplemented media by immunoblot method | 28233680 | |
| HCT116 | Function assay | 2 uM | 30 mins | Induction of AMPK phosphorylation at Thr-172 residue in human HCT116 cells at 2 uM after 30 mins in absence of glucose by immunoblot method | 28233680 | |
| U2OS | Antiviral assay | Antiviral activity against Chikungunya virus infected in human U2OS cells by RT-qPCR analysis, EC50=0.36μM | 28689975 | |||
| BHK-21 | Antiviral assay | Antiviral activity against Chikungunya virus 0611aTw infected in BHK-21 cells by RT-qPCR analysis, EC50=0.85μM | 28689975 | |||
| BHK-21 | Antiviral assay | Antiviral activity against Chikungunya virus 0810bTw infected in BHK-21 cells by RT-qPCR analysis, EC50=0.9μM | 28689975 | |||
| BHK-21 | Antiviral assay | Antiviral activity against Chikungunya virus infected in BHK-21 cells by RT-qPCR analysis, EC50=0.95μM | 28689975 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for DAOY cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SJ-GBM2 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for A673 cells | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-MC cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-37 cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB-EBc1 cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Saos-2 cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for SK-N-SH cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for NB1643 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for LAN-5 cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for BT-12 cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh18 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for OHS-50 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for RD cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for MG 63 (6-TG R) cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Primary screen for Rh41 cells | 29435139 | |||
| NB1643 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for NB1643 cells | 29435139 | |||
| A673 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for A673 cells) | 29435139 | |||
| SK-N-MC | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-MC cells | 29435139 | |||
| BT-12 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for BT-12 cells | 29435139 | |||
| LAN-5 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for LAN-5 cells | 29435139 | |||
| DAOY | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for DAOY cells | 29435139 | |||
| NB-EBc1 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for NB-EBc1 cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SJ-GBM2 cells | 29435139 | |||
| BT-37 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for BT-37 cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for TC32 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for MG 63 (6-TG R) cells | 29435139 | |||
| fibroblast cells | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for control Hh wild type fibroblast cells | 29435139 | |||
| U-2 OS | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for U-2 OS cells | 29435139 | |||
| Rh41 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh41 cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for RD cells | 29435139 | |||
| Rh18 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh18 cells | 29435139 | |||
| Rh30 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Rh30 cells | 29435139 | |||
| Saos-2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for Saos-2 cells | 29435139 | |||
| OHS-50 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for OHS-50 cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Confirmatory screen for SK-N-SH cells | 29435139 | |||
| SK-N-SH | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for SK-N-SH cells | 29435139 | |||
| SJ-GBM2 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for SJ-GBM2 cells | 29435139 | |||
| Daoy | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for Daoy cells | 29435139 | |||
| TC32 | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for TC32 cells | 29435139 | |||
| MG 63 (6-TG R) | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for MG 63 (6-TG R) cells | 29435139 | |||
| RD | qHTS assay | qHTS of pediatric cancer cell lines to identify multiple opportunities for drug repurposing: Orthogonal 3D viability screen for RD cells | 29435139 | |||
| SW948 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human SW948 cells after 72 hrs by colorimetric MTS assay, IC50=0.11μM | 30274939 | ||
| HT-29 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HT-29 cells after 72 hrs by colorimetric MTS assay, IC50=0.13μM | 30274939 | ||
| HCT116 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human HCT116 cells after 72 hrs by colorimetric MTS assay, IC50=0.41μM | 30274939 | ||
| HEK293 | Function assay | 6 hrs | Inhibition of Wnt3A-stimulated Wnt/beta-catenin pathway in HEK293 cells transfected with p8xTOPFlash, Renilla luciferase plasmid pRL-TK and pLKO.1 after 6 hrs by Dual topflash luciferase reporter assay, IC50=0.45μM | 30274939 | ||
| CRC240 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human CRC240 cells after 72 hrs by colorimetric MTS assay, IC50=0.89μM | 30274939 | ||
| SW480 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human SW480 cells after 72 hrs by colorimetric MTS assay, IC50=0.98μM | 30274939 | ||
| DLD1 | Antiproliferative assay | 72 hrs | Antiproliferative activity against human DLD1 cells after 72 hrs by colorimetric MTS assay, IC50=2.39μM | 30274939 | ||
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human HCT116 cells cytosol lysate harboring beta-catenin mutation assessed as reduction in Axin2 protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| HCT116 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human HCT116 cells cytosol lysate harboring beta-catenin mutation assessed as reduction in beta-catenin protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human SW480 cells cytosol lysate harboring APC mutation assessed as reduction in Axin2 protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| SW480 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human SW480 cells cytosol lysate harboring APC mutation assessed as reduction in beta-catenin protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| CRC240 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human CRC240 cells cytosol lysate assessed as reduction in Axin2 protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| CRC240 | Function assay | 5 uM | 18 hrs | Inhibition of Wnt/beta-catenin signaling pathway in human CRC240 cells cytosol lysate assessed as reduction in beta-catenin protein expression at 5 uM after 18 hrs by Western blot analysis | 30274939 | |
| CCD-841-CoN | Function assay | 1 uM | Stimulation of mitochondrial respiration in human CCD-841-CoN cells assessed as increase in oxygen consumption rate at 1 uM in presence of FCCP by polarographic analysis | 30274939 | ||
| CCD-841-CoN | Function assay | 0.1 to 10 uM | 2 hrs | Induction of AMPK phosphorylation in human CCD-841-CoN cells at 0.1 to 10 uM after 2 hrs by Western blot analysis | 30274939 | |
| CRC240 | Antitumor assay | 72 mg/kg | Antitumor activity against human CRC240 cells xenografted in NOD/SCID mouse at 72 mg/kg, po administered 11 days and measured on day 4, 8 during compound dosing and day 11 post last dose | 30274939 | ||
| U2OS | Function assay | 6 hrs | Induction of GFP-tagged Fzd1 (unknown origin) internalization expressed in human U2OS cells at 12.5 after 6 hrs by confocal microscopic method | 30274939 | ||
| A549 | Function assay | 6 hrs | Induction of apoptosis in human A549 cells assessed as increase in caspase 3/7 activity after 6 hrs by caspase glo 3/7 assay | 30371064 | ||
| Caco2 | Function assay | 6 hrs | Induction of apoptosis in human Caco2 cells assessed as increase in caspase 3/7 activity after 6 hrs by caspase glo 3/7 assay | 30371064 | ||
| AsPC1 | Function assay | 6 hrs | Induction of apoptosis in human AsPC1 cells assessed as increase in caspase 3/7 activity after 6 hrs by caspase glo 3/7 assay | 30371064 | ||
| HEK293 | Function assay | 8 hrs | Inhibition of Wnt3A-stimulated Wnt/beta-catenin pathway in HEK293 cells transfected with p8xTOPFlash, Renilla luciferase plasmid pRL-TK and pLKO.1 after 8 hrs by Dual topflash luciferase reporter gene assay, IC50=0.34μM | 30551901 | ||
| LN229 | Antiproliferative assay | 5 days | Antiproliferative activity against human LN229 cells after 5 days by AlamarBlue assay, IC50=0.3μM | 30583248 | ||
| T98G | Antiproliferative assay | 5 days | Antiproliferative activity against human T98G cells after 5 days by AlamarBlue assay, IC50=0.3μM | 30583248 | ||
| U87 | Antiproliferative assay | 5 days | Antiproliferative activity against human U87 cells after 5 days by AlamarBlue assay, IC50=0.3μM | 30583248 | ||
| U138MG | Antiproliferative assay | 5 days | Antiproliferative activity against human U138MG cells after 5 days by AlamarBlue assay, IC50=0.3μM | 30583248 | ||
| U373 | Antiproliferative assay | 5 days | Antiproliferative activity against human U373 cells after 5 days by AlamarBlue assay, IC50=0.3μM | 30583248 | ||
| HL60 | Cytotoxicity assay | 3 days | Cytotoxicity against human HL60 cells incubated for 3 days by CellTiter-Glo luminescent assay, IC50=0.3μM | 31253529 | ||
| KG1 | Cytotoxicity assay | 3 days | Cytotoxicity against human KG1 cells incubated for 3 days by CellTiter-Glo luminescent assay, IC50=0.36μM | 31253529 | ||
| Jurkat | Cytotoxicity assay | 3 days | Cytotoxicity against human Jurkat cells incubated for 3 days by CellTiter-Glo luminescent assay, IC50=0.4μM | 31253529 | ||
| NALM6 | Cytotoxicity assay | 3 days | Cytotoxicity against human NALM6 cells incubated for 3 days by CellTiter-Glo luminescent assay, IC50=0.62μM | 31253529 | ||
| HEK293 | Function assay | 90 mins | Inhibition of KIX-RLucN fused CBP (unknown origin) binding to KID-RLucC fused CREB (unknown origin) transfected in human HEK293 cells preincubated with compound 30 mins before before forskolin addition and measured after 90 mins in presence of coelenteraz, IC50=1.53μM | 31253529 | ||
| 293beta5 | Antiviral assay | 7 days | Antiviral activity against Human adenovirus 5 infected in 293beta5 cells assessed as inhibition of plaque formation incubated for 7 days by GFP reporter gene assay, IC50=0.6μM | 32045239 | ||
| A549 | Antiviral assay | 48 hrs | Antiviral activity against Human adenovirus 5 infected in human A549 cells assessed as inhibition of viral entry after 48 hrs by GFP reporter gene based assay, IC50=1.22μM | 32045239 | ||
| A549 | Cytotoxicity assay | 48 hrs | Cytotoxicity against human A549 cells assessed as reduction in cell viability after 48 hrs by alamar blue assay, CC50=22.9μM | 32045239 | ||
| Vero | Function assay | Vero cells viability qHTS for Zika virus inhibitors | 33229545 | |||
| Vero | Antiviral assay | 24 hr | Antiviral activity against SARS-CoV-2 (viral titer) measured by plaque assay in Vero cells at MOI 0.0125 after 24 hr, IC50=0.28μM | ChEMBL | ||
| skeletal myoblast cells | Cytotoxicity assay | 72 h | DNDI: Cytotoxicity in Vitro, 72 hour, in rat skeletal myoblast cells, IC50=2.3μM | ChEMBL | ||
| VERO-E6 | Function assay | 48 hrs | Determination of IC50 values for inhibition of SARS-CoV-2 induced cytotoxicity of VERO-E6 cells after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging, IC50=4.1μM | ChEMBL | ||
| Click to View More Cell Line Experimental Data | ||||||
Biological Activity
| Description | Niclosamide can inhibit DNA replication and inhibit STAT3 with IC50 of 0.7 μM in a cell-free assay. Niclosamide selectively inhibited the phosphorylation of STAT3 and had no obvious inhibition against the activation of other homologues (e.g., STAT1 and STAT5). | ||
|---|---|---|---|
| Targets |
|
| In vitro | ||||
| In vitro | Niclosamide (< 5 μM) dose dependently inhibits STAT3-mediated luciferase reporter activity with IC50 of 0.25 μM in HeLa cells. Niclosamide(< 2 μM) dose dependently inhibits the phosphorylation of STAT3 in Du145 cells. Niclosamide (1 μM) inhibits the EGF-induced nuclear translocation of STAT3 in Du145 cells. Niclosamide(< 2 μM) dose dependently inhibits the transcription of STAT3 downstream genes in Du145 cells. Niclosamide(< 10 μM) dose dependently induces G0/G1 arrest and apoptosis of Du145 cancer cells. [1] Niclosamide is able to inhibit SARS-CoV replication at a micromolar concentration in Vero E6 cells infected with SARS-CoV. [2] Niclosamide(< 7.5 μM) promotes Frizzled1 endocytosis, downregulates Dishevelled-2 protein, and inhibits Wnt3A-stimulated beta-catenin stabilization and LEF/TCF reporter activity in U2OS cells. [3] Niclosamide inhibits the TNF-induced NF-κB reporter activity in a dose- and time-dependent manner in U2OS cells. Niclosamide(125 nM) inhibits NF-κB activation induced by p65, IKKα, IKKβ, IKKγ, and TAK1 in U2OS cells. Niclosamide(< 500 nM) completely block the time- and dose-dependent TNFα-induced alteration of the NF-κB–DNA complex in HL-60 cells. Niclosamide(< 10 nM) inhibits constitutive NF-κB activation in U266 cells. Niclosamide inhibits TNF-induced degradation of IκBα and relocation of p65 in a dose- and time-dependent manner in HL-60, Molm13, or AML primary cells. Niclosamide(500 nM) decreases TNF-induced NF-κB–dependent gene products involved in cell survival in HL-60 cells. Niclosamide dose dependently inhibits the growth and induces robust apoptosis of AML cells associated with decreased Mcl-1 and XIAP levels and increased intracellular ROS levels. [4] | |||
|---|---|---|---|---|
| Kinase Assay | Protein Kinase profiling assay | |||
| Assay for 22 different proteins kinases is carried out by ProQinase Gmbh. All of the protein kinases are expressed either in Sf9 insect cells or in E.coli as recombinant GST-fusion proteins or His-tagged proteins. Protein kinases are purified by affinity chromatography using either GSH-agarose or Ni_NTH-agarose. A radiometric protein kinase assay is used for measuring the kinase activity of the 22 protein kinases. Briefly, for each protein kinase, 50 μL reaction cocktail containing 60 mM HEPES-NaOH, 3 mM MgCl2, 3 mM MnCl2, 3 μM Na-orthovanadate, 1.2 mM DTT, 50 μg/mL PEG20000, 1 μM [γ-33P]-ATP, Niclosamide, adequate amount of enzyme and its substrate. The PKC-alpha assay additionally contain 1 mM Cacl2, 4 mM EDTA, 5 μg/mL phosphatidylserine and 1 μg/mL 1, 2-Dioleyl-glycerol. The reaction cocktails are incubated at 37 °C for 60 minutes and stop with 50 μL 2% (v/v) H3PO4. Incorporation of 33Pi is determined with a microplate scintillation counter. The activities and the IC50 values are calculated using Quattro Workflow V2.28. | ||||
| Cell Research | Cell lines | Hela, A549, Du145, HT-29, A431, PC3 cells | ||
| Concentrations | 16 μM | |||
| Incubation Time | 72 hours | |||
| Method | Cells are plated in 96-well culture plates with cell density of 3-4 × 103 cells/well and treat with Niclosamide by adding 100 μL medium containing Niclosamide of various concentrations on the second day. After 72-hour's treatment, MTT is added to each well and incubated for additional 4-5 hours, and the absorbance is measured on a microplate reader at 570nm. Cell growth inhibition is evaluated as the ratio of the absorbance of the sample to that of the control. The results are representative of at least 3 independent experiments. |
|||
| Experimental Result Images | Methods | Biomarkers | Images | PMID |
| Western blot | β-catenin p-STAT5 / STAT5 / p-AKT / AKT / p-ERK / ERK p-BCR-ABL / BCR-ABL p-STAT3 / STAT3 / c-Myc / Survivin |
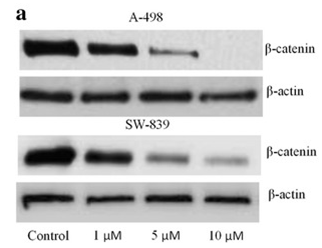
|
27652012 | |
| Growth inhibition assay | Cell viability |
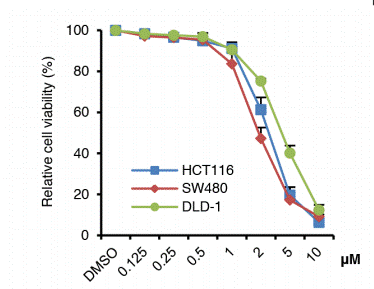
|
28418862 | |
| In Vivo | ||
| In vivo | Niclosamide(40 mg/kg/d, i.p.) inhibits growth of xenografted AML cells in nude mice bearing HL-60 xenograft tumors. [4] | |
|---|---|---|
| Animal Research | Animal Models | nude mice bearing HL-60 xenograft tumors. |
| Dosages | 40 mg/kg | |
| Administration | Intraperitoneal injection | |
| NCT Number | Recruitment | Conditions | Sponsor/Collaborators | Start Date | Phases |
|---|---|---|---|---|---|
| NCT05188170 | Recruiting | Acute Myeloid Leukemia (AML) |
Stanford University |
November 21 2022 | Phase 1 |
| NCT04436458 | Withdrawn | COVID |
First Wave BioPharma Inc. |
January 20 2022 | Phase 2 |
| NCT05168644 | Completed | Healthy |
TFF Pharmaceuticals Inc. |
November 14 2021 | Phase 1 |
| NCT04644705 | Completed | Healthy Volunteers |
Charité Research Organisation GmbH|Bayer |
November 2 2020 | Phase 1 |
| NCT04592835 | Unknown status | COVID-19 Patients |
Daewoong Pharmaceutical Co. LTD.|Novotech (Australia) Pty Limited |
October 19 2020 | Phase 1 |
| NCT04524052 | Unknown status | Healthy |
Daewoong Pharmaceutical Co. LTD. |
August 2020 | Phase 1 |
Chemical Information & Solubility
| Molecular Weight | 327.12 | Formula | C13H8Cl2N2O4 |
| CAS No. | 50-65-7 | SDF | Download Niclosamide SDF |
| Smiles | C1=CC(=C(C=C1[N+](=O)[O-])Cl)NC(=O)C2=C(C=CC(=C2)Cl)O | ||
| Storage (From the date of receipt) | |||
|
In vitro |
DMSO : 2 mg/mL ( (6.11 mM); Moisture-absorbing DMSO reduces solubility. Please use fresh DMSO.) Water : Insoluble Ethanol : Insoluble |
Molecular Weight Calculator |
|
In vivo Add solvents to the product individually and in order. |
In vivo Formulation Calculator |
||||
Preparing Stock Solutions
Molarity Calculator
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
% DMSO
%
% Tween 80
% ddH2O
%DMSO
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Tech Support
Answers to questions you may have can be found in the inhibitor handling instructions. Topics include how to prepare stock solutions, how to store inhibitors, and issues that need special attention for cell-based assays and animal experiments.
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
* Indicates a Required Field
Tags: buy Niclosamide | Niclosamide supplier | purchase Niclosamide | Niclosamide cost | Niclosamide manufacturer | order Niclosamide | Niclosamide distributor